Hang Wu
StreamVLA: Breaking the Reason-Act Cycle via Completion-State Gating
Feb 01, 2026Abstract:Long-horizon robotic manipulation requires bridging the gap between high-level planning (System 2) and low-level control (System 1). Current Vision-Language-Action (VLA) models often entangle these processes, performing redundant multimodal reasoning at every timestep, which leads to high latency and goal instability. To address this, we present StreamVLA, a dual-system architecture that unifies textual task decomposition, visual goal imagination, and continuous action generation within a single parameter-efficient backbone. We introduce a "Lock-and-Gated" mechanism to intelligently modulate computation: only when a sub-task transition is detected, the model triggers slow thinking to generate a textual instruction and imagines the specific visual completion state, rather than generic future frames. Crucially, this completion state serves as a time-invariant goal anchor, making the policy robust to execution speed variations. During steady execution, these high-level intents are locked to condition a Flow Matching action head, allowing the model to bypass expensive autoregressive decoding for 72% of timesteps. This hierarchical abstraction ensures sub-goal focus while significantly reducing inference latency. Extensive evaluations demonstrate that StreamVLA achieves state-of-the-art performance, with a 98.5% success rate on the LIBERO benchmark and robust recovery in real-world interference scenarios, achieving a 48% reduction in latency compared to full-reasoning baselines.
CamReasoner: Reinforcing Camera Movement Understanding via Structured Spatial Reasoning
Jan 30, 2026Abstract:Understanding camera dynamics is a fundamental pillar of video spatial intelligence. However, existing multimodal models predominantly treat this task as a black-box classification, often confusing physically distinct motions by relying on superficial visual patterns rather than geometric cues. We present CamReasoner, a framework that reformulates camera movement understanding as a structured inference process to bridge the gap between perception and cinematic logic. Our approach centers on the Observation-Thinking-Answer (O-T-A) paradigm, which compels the model to decode spatio-temporal cues such as trajectories and view frustums within an explicit reasoning block. To instill this capability, we construct a Large-scale Inference Trajectory Suite comprising 18k SFT reasoning chains and 38k RL feedback samples. Notably, we are the first to employ RL for logical alignment in this domain, ensuring motion inferences are grounded in physical geometry rather than contextual guesswork. By applying Reinforcement Learning to the Observation-Think-Answer (O-T-A) reasoning paradigm, CamReasoner effectively suppresses hallucinations and achieves state-of-the-art performance across multiple benchmarks.
PAS: A Training-Free Stabilizer for Temporal Encoding in Video LLMs
Nov 14, 2025Abstract:Video LLMs suffer from temporal inconsistency: small shifts in frame timing can flip attention and suppress relevant frames. We trace this instability to the common extension of Rotary Position Embeddings to video through multimodal RoPE. The induced inverse Fourier time kernel exhibits frame-scale ripples that multiply adjacent frames by different factors, which perturbs attention that should otherwise be governed by the raw query key inner product. We present Phase Aggregated Smoothing (PAS), a simple, training-free mechanism that applies small opposed phase offsets across heads and then aggregates their outputs. PAS preserves the per-head spectrum magnitude, while the aggregation effectively smooths the temporal kernel and reduces phase sensitivity without changing the positional encoding structure. Our analysis shows that the RoPE rotated logit can be approximated as a content dot product scaled by a time kernel; smoothing this kernel yields Lipschitz stability of attention to small temporal shifts; multi phase averaging attenuates high frequency ripples while preserving per-head spectra under Nyquist-valid sampling. Experiments on multiple video understanding benchmarks under matched token budgets show consistent improvements with negligible computational overhead. PAS provides a plug and play upgrade for robust temporal encoding in Video LLMs.
STAGE: A Stream-Centric Generative World Model for Long-Horizon Driving-Scene Simulation
Jun 16, 2025Abstract:The generation of temporally consistent, high-fidelity driving videos over extended horizons presents a fundamental challenge in autonomous driving world modeling. Existing approaches often suffer from error accumulation and feature misalignment due to inadequate decoupling of spatio-temporal dynamics and limited cross-frame feature propagation mechanisms. To address these limitations, we present STAGE (Streaming Temporal Attention Generative Engine), a novel auto-regressive framework that pioneers hierarchical feature coordination and multi-phase optimization for sustainable video synthesis. To achieve high-quality long-horizon driving video generation, we introduce Hierarchical Temporal Feature Transfer (HTFT) and a novel multi-stage training strategy. HTFT enhances temporal consistency between video frames throughout the video generation process by modeling the temporal and denoising process separately and transferring denoising features between frames. The multi-stage training strategy is to divide the training into three stages, through model decoupling and auto-regressive inference process simulation, thereby accelerating model convergence and reducing error accumulation. Experiments on the Nuscenes dataset show that STAGE has significantly surpassed existing methods in the long-horizon driving video generation task. In addition, we also explored STAGE's ability to generate unlimited-length driving videos. We generated 600 frames of high-quality driving videos on the Nuscenes dataset, which far exceeds the maximum length achievable by existing methods.
MedAgentGym: Training LLM Agents for Code-Based Medical Reasoning at Scale
Jun 04, 2025



Abstract:We introduce MedAgentGYM, the first publicly available training environment designed to enhance coding-based medical reasoning capabilities in large language model (LLM) agents. MedAgentGYM comprises 72,413 task instances across 129 categories derived from authentic real-world biomedical scenarios. Tasks are encapsulated within executable coding environments, each featuring detailed task descriptions, interactive feedback mechanisms, verifiable ground-truth annotations, and scalable training trajectory generation. Extensive benchmarking of over 30 LLMs reveals a notable performance disparity between commercial API-based models and open-source counterparts. Leveraging MedAgentGYM, Med-Copilot-7B achieves substantial performance gains through supervised fine-tuning (+36.44%) and continued reinforcement learning (+42.47%), emerging as an affordable and privacy-preserving alternative competitive with gpt-4o. By offering both a comprehensive benchmark and accessible, expandable training resources within unified execution environments, MedAgentGYM delivers an integrated platform to develop LLM-based coding assistants for advanced biomedical research and practice.
SpecRouter: Adaptive Routing for Multi-Level Speculative Decoding in Large Language Models
May 12, 2025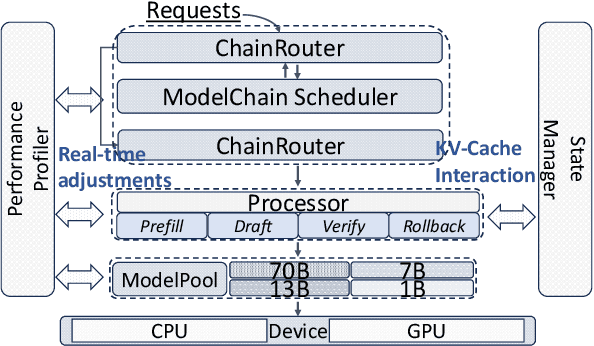
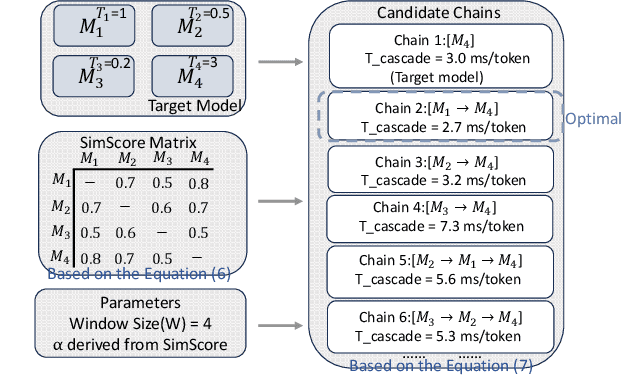
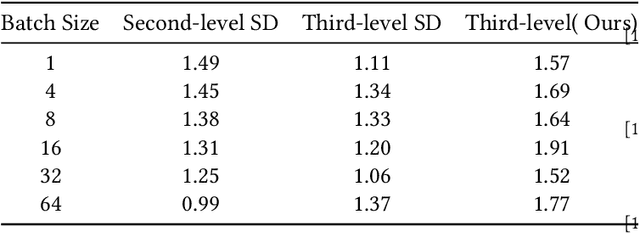
Abstract:Large Language Models (LLMs) present a critical trade-off between inference quality and computational cost: larger models offer superior capabilities but incur significant latency, while smaller models are faster but less powerful. Existing serving strategies often employ fixed model scales or static two-stage speculative decoding, failing to dynamically adapt to the varying complexities of user requests or fluctuations in system performance. This paper introduces \systemname{}, a novel framework that reimagines LLM inference as an adaptive routing problem solved through multi-level speculative decoding. \systemname{} dynamically constructs and optimizes inference "paths" (chains of models) based on real-time feedback, addressing the limitations of static approaches. Our contributions are threefold: (1) An \textbf{adaptive model chain scheduling} mechanism that leverages performance profiling (execution times) and predictive similarity metrics (derived from token distribution divergence) to continuously select the optimal sequence of draft and verifier models, minimizing predicted latency per generated token. (2) A \textbf{multi-level collaborative verification} framework where intermediate models within the selected chain can validate speculative tokens, reducing the verification burden on the final, most powerful target model. (3) A \textbf{synchronized state management} system providing efficient, consistent KV cache handling across heterogeneous models in the chain, including precise, low-overhead rollbacks tailored for asynchronous batch processing inherent in multi-level speculation. Preliminary experiments demonstrate the validity of our method.
BFS-Prover: Scalable Best-First Tree Search for LLM-based Automatic Theorem Proving
Feb 05, 2025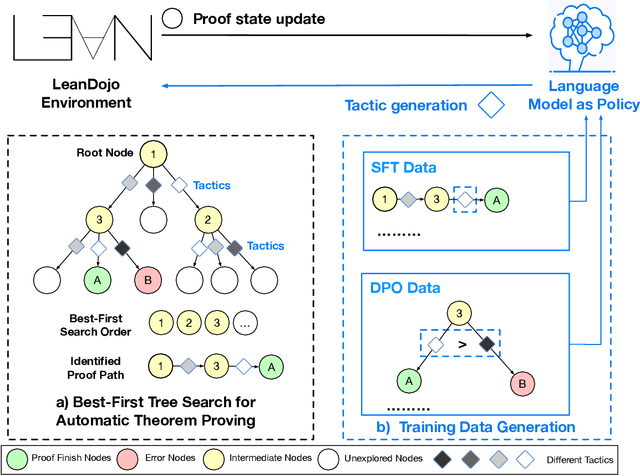
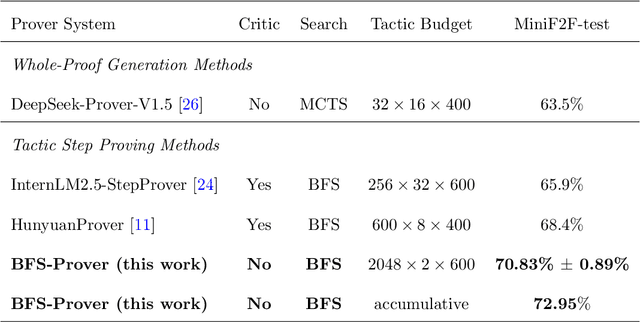
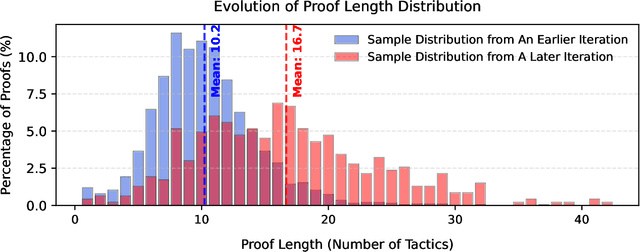
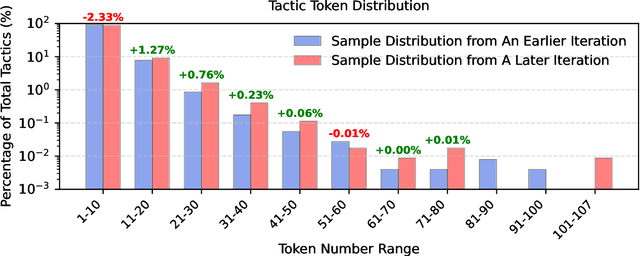
Abstract:Recent advancements in large language models (LLMs) have spurred growing interest in automatic theorem proving using Lean4, where effective tree search methods are crucial for navigating proof search spaces. While the existing approaches primarily rely on value functions and Monte Carlo Tree Search (MCTS), the potential of simpler methods like Best-First Search (BFS) remains underexplored. This paper investigates whether BFS can achieve competitive performance in large-scale theorem proving tasks. We present \texttt{BFS-Prover}, a scalable expert iteration framework, featuring three key innovations. First, we implement strategic data filtering at each expert iteration round, excluding problems solvable via beam search node expansion to focus on harder cases. Second, we improve the sample efficiency of BFS through Direct Preference Optimization (DPO) applied to state-tactic pairs automatically annotated with compiler error feedback, refining the LLM's policy to prioritize productive expansions. Third, we employ length normalization in BFS to encourage exploration of deeper proof paths. \texttt{BFS-Prover} achieves a score of $71.31$ on the MiniF2F test set and therefore challenges the perceived necessity of complex tree search methods, demonstrating that BFS can achieve competitive performance when properly scaled.
MapLocNet: Coarse-to-Fine Feature Registration for Visual Re-Localization in Navigation Maps
Jul 11, 2024



Abstract:Robust localization is the cornerstone of autonomous driving, especially in challenging urban environments where GPS signals suffer from multipath errors. Traditional localization approaches rely on high-definition (HD) maps, which consist of precisely annotated landmarks. However, building HD map is expensive and challenging to scale up. Given these limitations, leveraging navigation maps has emerged as a promising low-cost alternative for localization. Current approaches based on navigation maps can achieve highly accurate localization, but their complex matching strategies lead to unacceptable inference latency that fails to meet the real-time demands. To address these limitations, we propose a novel transformer-based neural re-localization method. Inspired by image registration, our approach performs a coarse-to-fine neural feature registration between navigation map and visual bird's-eye view features. Our method significantly outperforms the current state-of-the-art OrienterNet on both the nuScenes and Argoverse datasets, which is nearly 10%/20% localization accuracy and 30/16 FPS improvement on single-view and surround-view input settings, separately. We highlight that our research presents an HD-map-free localization method for autonomous driving, offering cost-effective, reliable, and scalable performance in challenging driving environments.
BLOS-BEV: Navigation Map Enhanced Lane Segmentation Network, Beyond Line of Sight
Jul 11, 2024



Abstract:Bird's-eye-view (BEV) representation is crucial for the perception function in autonomous driving tasks. It is difficult to balance the accuracy, efficiency and range of BEV representation. The existing works are restricted to a limited perception range within 50 meters. Extending the BEV representation range can greatly benefit downstream tasks such as topology reasoning, scene understanding, and planning by offering more comprehensive information and reaction time. The Standard-Definition (SD) navigation maps can provide a lightweight representation of road structure topology, characterized by ease of acquisition and low maintenance costs. An intuitive idea is to combine the close-range visual information from onboard cameras with the beyond line-of-sight (BLOS) environmental priors from SD maps to realize expanded perceptual capabilities. In this paper, we propose BLOS-BEV, a novel BEV segmentation model that incorporates SD maps for accurate beyond line-of-sight perception, up to 200m. Our approach is applicable to common BEV architectures and can achieve excellent results by incorporating information derived from SD maps. We explore various feature fusion schemes to effectively integrate the visual BEV representations and semantic features from the SD map, aiming to leverage the complementary information from both sources optimally. Extensive experiments demonstrate that our approach achieves state-of-the-art performance in BEV segmentation on nuScenes and Argoverse benchmark. Through multi-modal inputs, BEV segmentation is significantly enhanced at close ranges below 50m, while also demonstrating superior performance in long-range scenarios, surpassing other methods by over 20% mIoU at distances ranging from 50-200m.
MedAdapter: Efficient Test-Time Adaptation of Large Language Models towards Medical Reasoning
May 05, 2024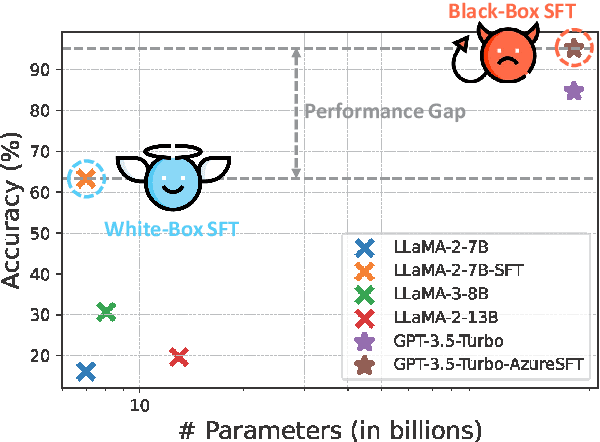
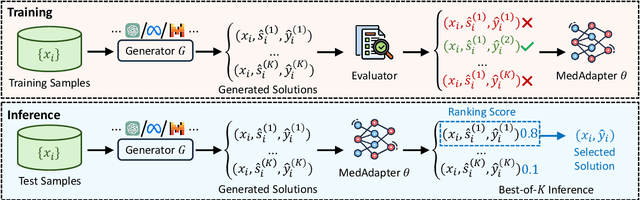
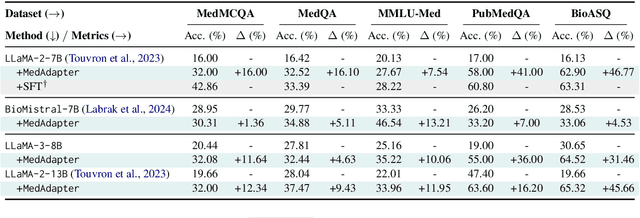
Abstract:Despite their improved capabilities in generation and reasoning, adapting large language models (LLMs) to the biomedical domain remains challenging due to their immense size and corporate privacy. In this work, we propose MedAdapter, a unified post-hoc adapter for test-time adaptation of LLMs towards biomedical applications. Instead of fine-tuning the entire LLM, MedAdapter effectively adapts the original model by fine-tuning only a small BERT-sized adapter to rank candidate solutions generated by LLMs. Experiments demonstrate that MedAdapter effectively adapts both white-box and black-box LLMs in biomedical reasoning, achieving average performance improvements of 25.48% and 11.31%, respectively, without requiring extensive computational resources or sharing data with third parties. MedAdapter also yields superior performance when combined with train-time adaptation, highlighting a flexible and complementary solution to existing adaptation methods. Faced with the challenges of balancing model performance, computational resources, and data privacy, MedAdapter provides an efficient, privacy-preserving, cost-effective, and transparent solution for adapting LLMs to the biomedical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge