Joyce Ho
PromptLink: Leveraging Large Language Models for Cross-Source Biomedical Concept Linking
May 13, 2024



Abstract:Linking (aligning) biomedical concepts across diverse data sources enables various integrative analyses, but it is challenging due to the discrepancies in concept naming conventions. Various strategies have been developed to overcome this challenge, such as those based on string-matching rules, manually crafted thesauri, and machine learning models. However, these methods are constrained by limited prior biomedical knowledge and can hardly generalize beyond the limited amounts of rules, thesauri, or training samples. Recently, large language models (LLMs) have exhibited impressive results in diverse biomedical NLP tasks due to their unprecedentedly rich prior knowledge and strong zero-shot prediction abilities. However, LLMs suffer from issues including high costs, limited context length, and unreliable predictions. In this research, we propose PromptLink, a novel biomedical concept linking framework that leverages LLMs. It first employs a biomedical-specialized pre-trained language model to generate candidate concepts that can fit in the LLM context windows. Then it utilizes an LLM to link concepts through two-stage prompts, where the first-stage prompt aims to elicit the biomedical prior knowledge from the LLM for the concept linking task and the second-stage prompt enforces the LLM to reflect on its own predictions to further enhance their reliability. Empirical results on the concept linking task between two EHR datasets and an external biomedical KG demonstrate the effectiveness of PromptLink. Furthermore, PromptLink is a generic framework without reliance on additional prior knowledge, context, or training data, making it well-suited for concept linking across various types of data sources. The source code is available at https://github.com/constantjxyz/PromptLink.
EHRAgent: Code Empowers Large Language Models for Complex Tabular Reasoning on Electronic Health Records
Jan 13, 2024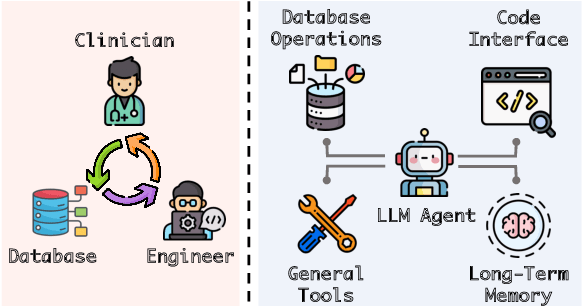
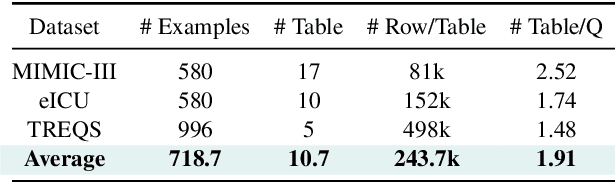
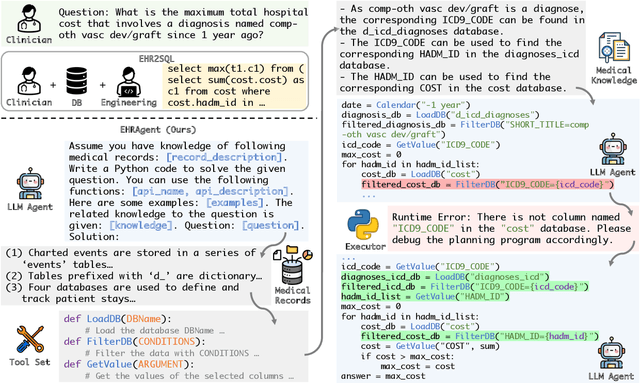
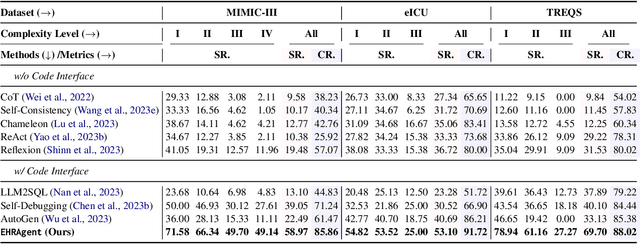
Abstract:Large language models (LLMs) have demonstrated exceptional capabilities in planning and tool utilization as autonomous agents, but few have been developed for medical problem-solving. We propose EHRAgent1, an LLM agent empowered with a code interface, to autonomously generate and execute code for complex clinical tasks within electronic health records (EHRs). First, we formulate an EHR question-answering task into a tool-use planning process, efficiently decomposing a complicated task into a sequence of manageable actions. By integrating interactive coding and execution feedback, EHRAgent learns from error messages and improves the originally generated code through iterations. Furthermore, we enhance the LLM agent by incorporating long-term memory, which allows EHRAgent to effectively select and build upon the most relevant successful cases from past experiences. Experiments on two real-world EHR datasets show that EHRAgent outperforms the strongest LLM agent baseline by 36.48% and 12.41%, respectively. EHRAgent leverages the emerging few-shot learning capabilities of LLMs, enabling autonomous code generation and execution to tackle complex clinical tasks with minimal demonstrations.
Knowledge-Infused Prompting: Assessing and Advancing Clinical Text Data Generation with Large Language Models
Nov 01, 2023



Abstract:Clinical natural language processing requires methods that can address domain-specific challenges, such as complex medical terminology and clinical contexts. Recently, large language models (LLMs) have shown promise in this domain. Yet, their direct deployment can lead to privacy issues and are constrained by resources. To address this challenge, we delve into synthetic clinical text generation using LLMs for clinical NLP tasks. We propose an innovative, resource-efficient approach, ClinGen, which infuses knowledge into the process. Our model involves clinical knowledge extraction and context-informed LLM prompting. Both clinical topics and writing styles are drawn from external domain-specific knowledge graphs and LLMs to guide data generation. Our extensive empirical study across 7 clinical NLP tasks and 16 datasets reveals that ClinGen consistently enhances performance across various tasks, effectively aligning the distribution of real datasets and significantly enriching the diversity of generated training instances. We will publish our code and all the generated data in \url{https://github.com/ritaranx/ClinGen}.
A Survey on Knowledge Graphs for Healthcare: Resources, Applications, and Promises
Jun 07, 2023



Abstract:Healthcare knowledge graphs (HKGs) have emerged as a promising tool for organizing medical knowledge in a structured and interpretable way, which provides a comprehensive view of medical concepts and their relationships. However, challenges such as data heterogeneity and limited coverage remain, emphasizing the need for further research in the field of HKGs. This survey paper serves as the first comprehensive overview of HKGs. We summarize the pipeline and key techniques for HKG construction (i.e., from scratch and through integration), as well as the common utilization approaches (i.e., model-free and model-based). To provide researchers with valuable resources, we organize existing HKGs (The resource is available at https://github.com/lujiaying/Awesome-HealthCare-KnowledgeBase) based on the data types they capture and application domains, supplemented with pertinent statistical information. In the application section, we delve into the transformative impact of HKGs across various healthcare domains, spanning from fine-grained basic science research to high-level clinical decision support. Lastly, we shed light on the opportunities for creating comprehensive and accurate HKGs in the era of large language models, presenting the potential to revolutionize healthcare delivery and enhance the interpretability and reliability of clinical prediction.
Neighborhood-Regularized Self-Training for Learning with Few Labels
Jan 10, 2023



Abstract:Training deep neural networks (DNNs) with limited supervision has been a popular research topic as it can significantly alleviate the annotation burden. Self-training has been successfully applied in semi-supervised learning tasks, but one drawback of self-training is that it is vulnerable to the label noise from incorrect pseudo labels. Inspired by the fact that samples with similar labels tend to share similar representations, we develop a neighborhood-based sample selection approach to tackle the issue of noisy pseudo labels. We further stabilize self-training via aggregating the predictions from different rounds during sample selection. Experiments on eight tasks show that our proposed method outperforms the strongest self-training baseline with 1.83% and 2.51% performance gain for text and graph datasets on average. Our further analysis demonstrates that our proposed data selection strategy reduces the noise of pseudo labels by 36.8% and saves 57.3% of the time when compared with the best baseline. Our code and appendices will be uploaded to https://github.com/ritaranx/NeST.
TASTE: Temporal and Static Tensor Factorization for Phenotyping Electronic Health Records
Nov 13, 2019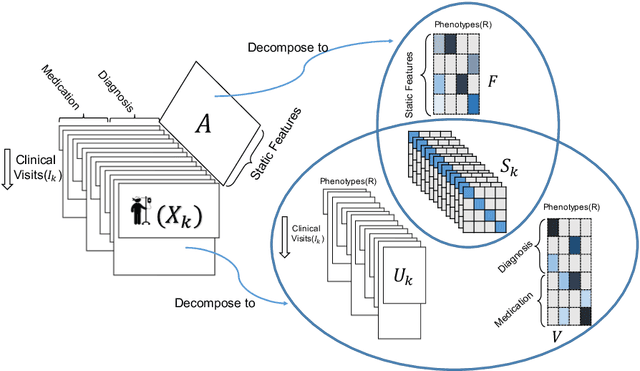
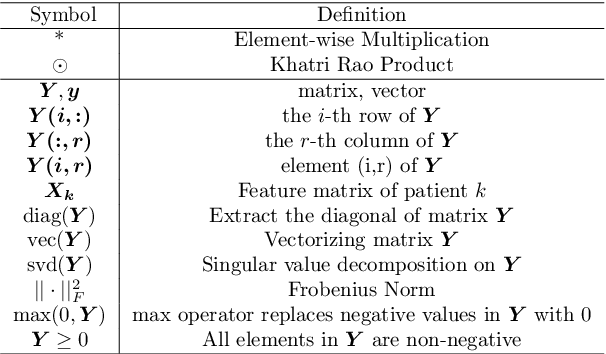
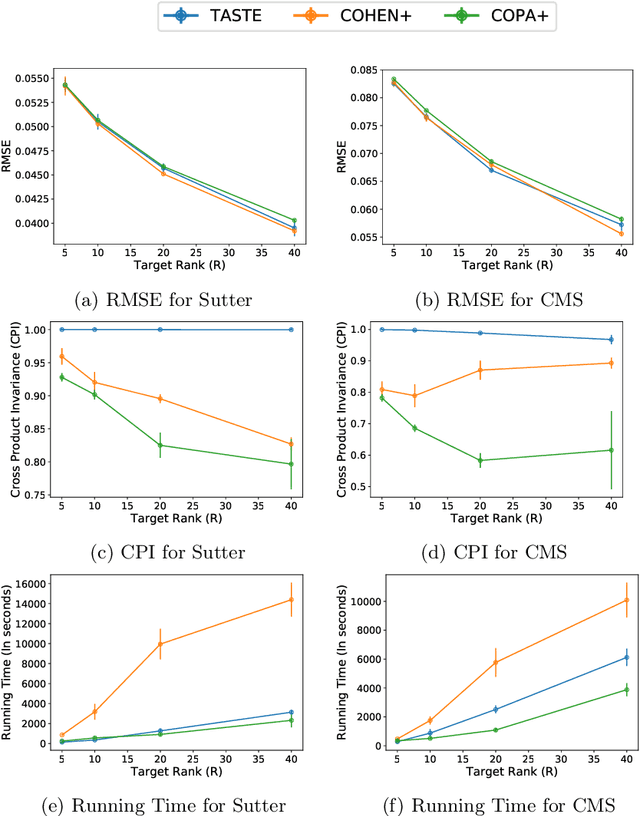
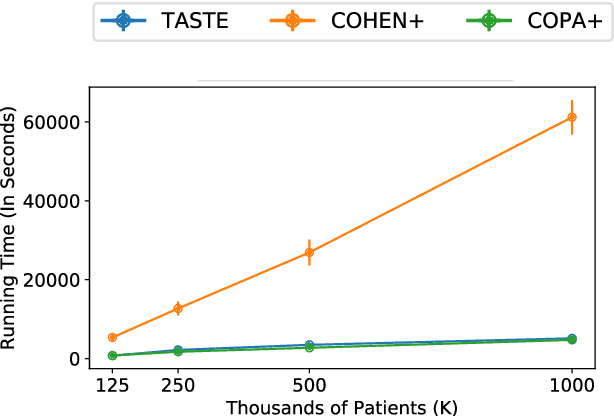
Abstract:Phenotyping electronic health records (EHR) focuses on defining meaningful patient groups (e.g., heart failure group and diabetes group) and identifying the temporal evolution of patients in those groups. Tensor factorization has been an effective tool for phenotyping. Most of the existing works assume either a static patient representation with aggregate data or only model temporal data. However, real EHR data contain both temporal (e.g., longitudinal clinical visits) and static information (e.g., patient demographics), which are difficult to model simultaneously. In this paper, we propose Temporal And Static TEnsor factorization (TASTE) that jointly models both static and temporal information to extract phenotypes. TASTE combines the PARAFAC2 model with non-negative matrix factorization to model a temporal and a static tensor. To fit the proposed model, we transform the original problem into simpler ones which are optimally solved in an alternating fashion. For each of the sub-problems, our proposed mathematical reformulations lead to efficient sub-problem solvers. Comprehensive experiments on large EHR data from a heart failure (HF) study confirmed that TASTE is up to 14x faster than several baselines and the resulting phenotypes were confirmed to be clinically meaningful by a cardiologist. Using 80 phenotypes extracted by TASTE, a simple logistic regression can achieve the same level of area under the curve (AUC) for HF prediction compared to a deep learning model using recurrent neural networks (RNN) with 345 features.
COPA: Constrained PARAFAC2 for Sparse & Large Datasets
Aug 27, 2018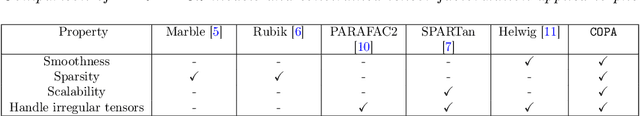
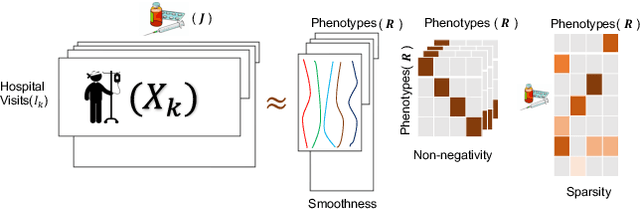
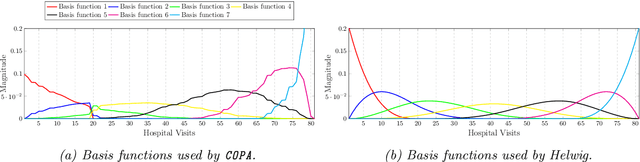
Abstract:PARAFAC2 has demonstrated success in modeling irregular tensors, where the tensor dimensions vary across one of the modes. An example scenario is modeling treatments across a set of patients with the varying number of medical encounters over time. Despite recent improvements on unconstrained PARAFAC2, its model factors are usually dense and sensitive to noise which limits their interpretability. As a result, the following open challenges remain: a) various modeling constraints, such as temporal smoothness, sparsity and non-negativity, are needed to be imposed for interpretable temporal modeling and b) a scalable approach is required to support those constraints efficiently for large datasets. To tackle these challenges, we propose a {\it CO}nstrained {\it PA}RAFAC2 (COPA) method, which carefully incorporates optimization constraints such as temporal smoothness, sparsity, and non-negativity in the resulting factors. To efficiently support all those constraints, COPA adopts a hybrid optimization framework using alternating optimization and alternating direction method of multiplier (AO-ADMM). As evaluated on large electronic health record (EHR) datasets with hundreds of thousands of patients, COPA achieves significant speedups (up to 36 times faster) over prior PARAFAC2 approaches that only attempt to handle a subset of the constraints that COPA enables. Overall, our method outperforms all the baselines attempting to handle a subset of the constraints in terms of speed, while achieving the same level of accuracy. Through a case study on temporal phenotyping of medically complex children, we demonstrate how the constraints imposed by COPA reveal concise phenotypes and meaningful temporal profiles of patients. The clinical interpretation of both the phenotypes and the temporal profiles was confirmed by a medical expert.
ACDC: $α$-Carving Decision Chain for Risk Stratification
Jun 16, 2016

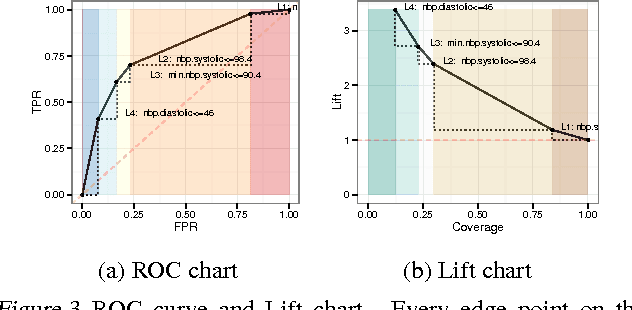

Abstract:In many healthcare settings, intuitive decision rules for risk stratification can help effective hospital resource allocation. This paper introduces a novel variant of decision tree algorithms that produces a chain of decisions, not a general tree. Our algorithm, $\alpha$-Carving Decision Chain (ACDC), sequentially carves out "pure" subsets of the majority class examples. The resulting chain of decision rules yields a pure subset of the minority class examples. Our approach is particularly effective in exploring large and class-imbalanced health datasets. Moreover, ACDC provides an interactive interpretation in conjunction with visual performance metrics such as Receiver Operating Characteristics curve and Lift chart.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge