Xuan Kan
Parameter-Efficient Transfer Learning under Federated Learning for Automatic Speech Recognition
Aug 19, 2024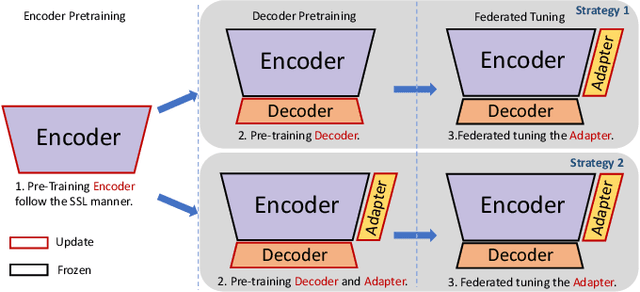
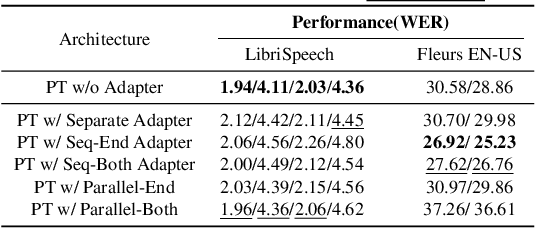
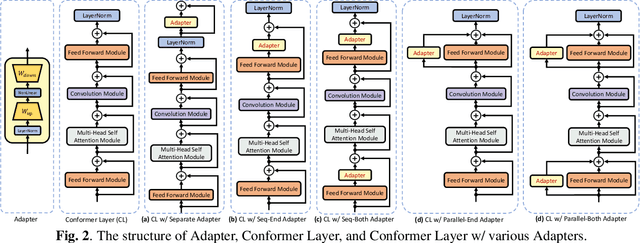
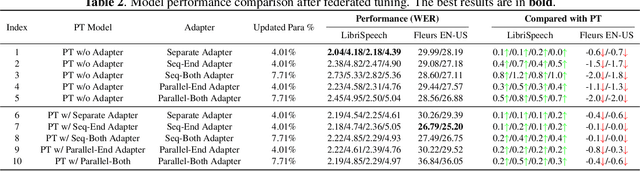
Abstract:This work explores the challenge of enhancing Automatic Speech Recognition (ASR) model performance across various user-specific domains while preserving user data privacy. We employ federated learning and parameter-efficient domain adaptation methods to solve the (1) massive data requirement of ASR models from user-specific scenarios and (2) the substantial communication cost between servers and clients during federated learning. We demonstrate that when equipped with proper adapters, ASR models under federated tuning can achieve similar performance compared with centralized tuning ones, thus providing a potential direction for future privacy-preserved ASR services. Besides, we investigate the efficiency of different adapters and adapter incorporation strategies under the federated learning setting.
BrainODE: Dynamic Brain Signal Analysis via Graph-Aided Neural Ordinary Differential Equations
Apr 30, 2024



Abstract:Brain network analysis is vital for understanding the neural interactions regarding brain structures and functions, and identifying potential biomarkers for clinical phenotypes. However, widely used brain signals such as Blood Oxygen Level Dependent (BOLD) time series generated from functional Magnetic Resonance Imaging (fMRI) often manifest three challenges: (1) missing values, (2) irregular samples, and (3) sampling misalignment, due to instrumental limitations, impacting downstream brain network analysis and clinical outcome predictions. In this work, we propose a novel model called BrainODE to achieve continuous modeling of dynamic brain signals using Ordinary Differential Equations (ODE). By learning latent initial values and neural ODE functions from irregular time series, BrainODE effectively reconstructs brain signals at any time point, mitigating the aforementioned three data challenges of brain signals altogether. Comprehensive experimental results on real-world neuroimaging datasets demonstrate the superior performance of BrainODE and its capability of addressing the three data challenges.
Knowledge-Infused Prompting: Assessing and Advancing Clinical Text Data Generation with Large Language Models
Nov 01, 2023



Abstract:Clinical natural language processing requires methods that can address domain-specific challenges, such as complex medical terminology and clinical contexts. Recently, large language models (LLMs) have shown promise in this domain. Yet, their direct deployment can lead to privacy issues and are constrained by resources. To address this challenge, we delve into synthetic clinical text generation using LLMs for clinical NLP tasks. We propose an innovative, resource-efficient approach, ClinGen, which infuses knowledge into the process. Our model involves clinical knowledge extraction and context-informed LLM prompting. Both clinical topics and writing styles are drawn from external domain-specific knowledge graphs and LLMs to guide data generation. Our extensive empirical study across 7 clinical NLP tasks and 16 datasets reveals that ClinGen consistently enhances performance across various tasks, effectively aligning the distribution of real datasets and significantly enriching the diversity of generated training instances. We will publish our code and all the generated data in \url{https://github.com/ritaranx/ClinGen}.
Open Visual Knowledge Extraction via Relation-Oriented Multimodality Model Prompting
Oct 28, 2023



Abstract:Images contain rich relational knowledge that can help machines understand the world. Existing methods on visual knowledge extraction often rely on the pre-defined format (e.g., sub-verb-obj tuples) or vocabulary (e.g., relation types), restricting the expressiveness of the extracted knowledge. In this work, we take a first exploration to a new paradigm of open visual knowledge extraction. To achieve this, we present OpenVik which consists of an open relational region detector to detect regions potentially containing relational knowledge and a visual knowledge generator that generates format-free knowledge by prompting the large multimodality model with the detected region of interest. We also explore two data enhancement techniques for diversifying the generated format-free visual knowledge. Extensive knowledge quality evaluations highlight the correctness and uniqueness of the extracted open visual knowledge by OpenVik. Moreover, integrating our extracted knowledge across various visual reasoning applications shows consistent improvements, indicating the real-world applicability of OpenVik.
Dynamic Brain Transformer with Multi-level Attention for Functional Brain Network Analysis
Sep 05, 2023Abstract:Recent neuroimaging studies have highlighted the importance of network-centric brain analysis, particularly with functional magnetic resonance imaging. The emergence of Deep Neural Networks has fostered a substantial interest in predicting clinical outcomes and categorizing individuals based on brain networks. However, the conventional approach involving static brain network analysis offers limited potential in capturing the dynamism of brain function. Although recent studies have attempted to harness dynamic brain networks, their high dimensionality and complexity present substantial challenges. This paper proposes a novel methodology, Dynamic bRAin Transformer (DART), which combines static and dynamic brain networks for more effective and nuanced brain function analysis. Our model uses the static brain network as a baseline, integrating dynamic brain networks to enhance performance against traditional methods. We innovatively employ attention mechanisms, enhancing model explainability and exploiting the dynamic brain network's temporal variations. The proposed approach offers a robust solution to the low signal-to-noise ratio of blood-oxygen-level-dependent signals, a recurring issue in direct DNN modeling. It also provides valuable insights into which brain circuits or dynamic networks contribute more to final predictions. As such, DRAT shows a promising direction in neuroimaging studies, contributing to the comprehensive understanding of brain organization and the role of neural circuits.
A Survey on Knowledge Graphs for Healthcare: Resources, Applications, and Promises
Jun 07, 2023



Abstract:Healthcare knowledge graphs (HKGs) have emerged as a promising tool for organizing medical knowledge in a structured and interpretable way, which provides a comprehensive view of medical concepts and their relationships. However, challenges such as data heterogeneity and limited coverage remain, emphasizing the need for further research in the field of HKGs. This survey paper serves as the first comprehensive overview of HKGs. We summarize the pipeline and key techniques for HKG construction (i.e., from scratch and through integration), as well as the common utilization approaches (i.e., model-free and model-based). To provide researchers with valuable resources, we organize existing HKGs (The resource is available at https://github.com/lujiaying/Awesome-HealthCare-KnowledgeBase) based on the data types they capture and application domains, supplemented with pertinent statistical information. In the application section, we delve into the transformative impact of HKGs across various healthcare domains, spanning from fine-grained basic science research to high-level clinical decision support. Lastly, we shed light on the opportunities for creating comprehensive and accurate HKGs in the era of large language models, presenting the potential to revolutionize healthcare delivery and enhance the interpretability and reliability of clinical prediction.
R-Mixup: Riemannian Mixup for Biological Networks
Jun 05, 2023



Abstract:Biological networks are commonly used in biomedical and healthcare domains to effectively model the structure of complex biological systems with interactions linking biological entities. However, due to their characteristics of high dimensionality and low sample size, directly applying deep learning models on biological networks usually faces severe overfitting. In this work, we propose R-MIXUP, a Mixup-based data augmentation technique that suits the symmetric positive definite (SPD) property of adjacency matrices from biological networks with optimized training efficiency. The interpolation process in R-MIXUP leverages the log-Euclidean distance metrics from the Riemannian manifold, effectively addressing the swelling effect and arbitrarily incorrect label issues of vanilla Mixup. We demonstrate the effectiveness of R-MIXUP with five real-world biological network datasets on both regression and classification tasks. Besides, we derive a commonly ignored necessary condition for identifying the SPD matrices of biological networks and empirically study its influence on the model performance. The code implementation can be found in Appendix E.
Transformer-Based Hierarchical Clustering for Brain Network Analysis
May 06, 2023



Abstract:Brain networks, graphical models such as those constructed from MRI, have been widely used in pathological prediction and analysis of brain functions. Within the complex brain system, differences in neuronal connection strengths parcellate the brain into various functional modules (network communities), which are critical for brain analysis. However, identifying such communities within the brain has been a nontrivial issue due to the complexity of neuronal interactions. In this work, we propose a novel interpretable transformer-based model for joint hierarchical cluster identification and brain network classification. Extensive experimental results on real-world brain network datasets show that with the help of hierarchical clustering, the model achieves increased accuracy and reduced runtime complexity while providing plausible insight into the functional organization of brain regions. The implementation is available at https://github.com/DDVD233/THC.
Neighborhood-Regularized Self-Training for Learning with Few Labels
Jan 10, 2023



Abstract:Training deep neural networks (DNNs) with limited supervision has been a popular research topic as it can significantly alleviate the annotation burden. Self-training has been successfully applied in semi-supervised learning tasks, but one drawback of self-training is that it is vulnerable to the label noise from incorrect pseudo labels. Inspired by the fact that samples with similar labels tend to share similar representations, we develop a neighborhood-based sample selection approach to tackle the issue of noisy pseudo labels. We further stabilize self-training via aggregating the predictions from different rounds during sample selection. Experiments on eight tasks show that our proposed method outperforms the strongest self-training baseline with 1.83% and 2.51% performance gain for text and graph datasets on average. Our further analysis demonstrates that our proposed data selection strategy reduces the noise of pseudo labels by 36.8% and saves 57.3% of the time when compared with the best baseline. Our code and appendices will be uploaded to https://github.com/ritaranx/NeST.
Learning Task-Aware Effective Brain Connectivity for fMRI Analysis with Graph Neural Networks
Nov 01, 2022



Abstract:Functional magnetic resonance imaging (fMRI) has become one of the most common imaging modalities for brain function analysis. Recently, graph neural networks (GNN) have been adopted for fMRI analysis with superior performance. Unfortunately, traditional functional brain networks are mainly constructed based on similarities among region of interests (ROI), which are noisy and agnostic to the downstream prediction tasks and can lead to inferior results for GNN-based models. To better adapt GNNs for fMRI analysis, we propose TBDS, an end-to-end framework based on \underline{T}ask-aware \underline{B}rain connectivity \underline{D}AG (short for Directed Acyclic Graph) \underline{S}tructure generation for fMRI analysis. The key component of TBDS is the brain network generator which adopts a DAG learning approach to transform the raw time-series into task-aware brain connectivities. Besides, we design an additional contrastive regularization to inject task-specific knowledge during the brain network generation process. Comprehensive experiments on two fMRI datasets, namely Adolescent Brain Cognitive Development (ABCD) and Philadelphia Neuroimaging Cohort (PNC) datasets demonstrate the efficacy of TBDS. In addition, the generated brain networks also highlight the prediction-related brain regions and thus provide unique interpretations of the prediction results. Our implementation will be published to https://github.com/yueyu1030/TBDS upon acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge