Yizhou Sun
FlexLLM: Composable HLS Library for Flexible Hybrid LLM Accelerator Design
Jan 22, 2026Abstract:We present FlexLLM, a composable High-Level Synthesis (HLS) library for rapid development of domain-specific LLM accelerators. FlexLLM exposes key architectural degrees of freedom for stage-customized inference, enabling hybrid designs that tailor temporal reuse and spatial dataflow differently for prefill and decode, and provides a comprehensive quantization suite to support accurate low-bit deployment. Using FlexLLM, we build a complete inference system for the Llama-3.2 1B model in under two months with only 1K lines of code. The system includes: (1) a stage-customized accelerator with hardware-efficient quantization (12.68 WikiText-2 PPL) surpassing SpinQuant baseline, and (2) a Hierarchical Memory Transformer (HMT) plug-in for efficient long-context processing. On the AMD U280 FPGA at 16nm, the accelerator achieves 1.29$\times$ end-to-end speedup, 1.64$\times$ higher decode throughput, and 3.14$\times$ better energy efficiency than an NVIDIA A100 GPU (7nm) running BF16 inference; projected results on the V80 FPGA at 7nm reach 4.71$\times$, 6.55$\times$, and 4.13$\times$, respectively. In long-context scenarios, integrating the HMT plug-in reduces prefill latency by 23.23$\times$ and extends the context window by 64$\times$, delivering 1.10$\times$/4.86$\times$ lower end-to-end latency and 5.21$\times$/6.27$\times$ higher energy efficiency on the U280/V80 compared to the A100 baseline. FlexLLM thus bridges algorithmic innovation in LLM inference and high-performance accelerators with minimal manual effort.
Report for NSF Workshop on AI for Electronic Design Automation
Jan 20, 2026Abstract:This report distills the discussions and recommendations from the NSF Workshop on AI for Electronic Design Automation (EDA), held on December 10, 2024 in Vancouver alongside NeurIPS 2024. Bringing together experts across machine learning and EDA, the workshop examined how AI-spanning large language models (LLMs), graph neural networks (GNNs), reinforcement learning (RL), neurosymbolic methods, etc.-can facilitate EDA and shorten design turnaround. The workshop includes four themes: (1) AI for physical synthesis and design for manufacturing (DFM), discussing challenges in physical manufacturing process and potential AI applications; (2) AI for high-level and logic-level synthesis (HLS/LLS), covering pragma insertion, program transformation, RTL code generation, etc.; (3) AI toolbox for optimization and design, discussing frontier AI developments that could potentially be applied to EDA tasks; and (4) AI for test and verification, including LLM-assisted verification tools, ML-augmented SAT solving, security/reliability challenges, etc. The report recommends NSF to foster AI/EDA collaboration, invest in foundational AI for EDA, develop robust data infrastructures, promote scalable compute infrastructure, and invest in workforce development to democratize hardware design and enable next-generation hardware systems. The workshop information can be found on the website https://ai4eda-workshop.github.io/.
Simulator and Experience Enhanced Diffusion Model for Comprehensive ECG Generation
Nov 13, 2025Abstract:Cardiovascular disease (CVD) is a leading cause of mortality worldwide. Electrocardiograms (ECGs) are the most widely used non-invasive tool for cardiac assessment, yet large, well-annotated ECG corpora are scarce due to cost, privacy, and workflow constraints. Generating ECGs can be beneficial for the mechanistic understanding of cardiac electrical activity, enable the construction of large, heterogeneous, and unbiased datasets, and facilitate privacy-preserving data sharing. Generating realistic ECG signals from clinical context is important yet underexplored. Recent work has leveraged diffusion models for text-to-ECG generation, but two challenges remain: (i) existing methods often overlook the physiological simulator knowledge of cardiac activity; and (ii) they ignore broader, experience-based clinical knowledge grounded in real-world practice. To address these gaps, we propose SE-Diff, a novel physiological simulator and experience enhanced diffusion model for comprehensive ECG generation. SE-Diff integrates a lightweight ordinary differential equation (ODE)-based ECG simulator into the diffusion process via a beat decoder and simulator-consistent constraints, injecting mechanistic priors that promote physiologically plausible waveforms. In parallel, we design an LLM-powered experience retrieval-augmented strategy to inject clinical knowledge, providing more guidance for ECG generation. Extensive experiments on real-world ECG datasets demonstrate that SE-Diff improves both signal fidelity and text-ECG semantic alignment over baselines, proving its superiority for text-to-ECG generation. We further show that the simulator-based and experience-based knowledge also benefit downstream ECG classification.
Conditional Neural ODE for Longitudinal Parkinson's Disease Progression Forecasting
Nov 06, 2025Abstract:Parkinson's disease (PD) shows heterogeneous, evolving brain-morphometry patterns. Modeling these longitudinal trajectories enables mechanistic insight, treatment development, and individualized 'digital-twin' forecasting. However, existing methods usually adopt recurrent neural networks and transformer architectures, which rely on discrete, regularly sampled data while struggling to handle irregular and sparse magnetic resonance imaging (MRI) in PD cohorts. Moreover, these methods have difficulty capturing individual heterogeneity including variations in disease onset, progression rate, and symptom severity, which is a hallmark of PD. To address these challenges, we propose CNODE (Conditional Neural ODE), a novel framework for continuous, individualized PD progression forecasting. The core of CNODE is to model morphological brain changes as continuous temporal processes using a neural ODE model. In addition, we jointly learn patient-specific initial time and progress speed to align individual trajectories into a shared progression trajectory. We validate CNODE on the Parkinson's Progression Markers Initiative (PPMI) dataset. Experimental results show that our method outperforms state-of-the-art baselines in forecasting longitudinal PD progression.
Dynamic Generation of Multi-LLM Agents Communication Topologies with Graph Diffusion Models
Oct 09, 2025Abstract:The efficiency of multi-agent systems driven by large language models (LLMs) largely hinges on their communication topology. However, designing an optimal topology is a non-trivial challenge, as it requires balancing competing objectives such as task performance, communication cost, and robustness. Existing frameworks often rely on static or hand-crafted topologies, which inherently fail to adapt to diverse task requirements, leading to either excessive token consumption for simple problems or performance bottlenecks for complex ones. To address this challenge, we introduce a novel generative framework called \textit{Guided Topology Diffusion (GTD)}. Inspired by conditional discrete graph diffusion models, GTD formulates topology synthesis as an iterative construction process. At each step, the generation is steered by a lightweight proxy model that predicts multi-objective rewards (e.g., accuracy, utility, cost), enabling real-time, gradient-free optimization towards task-adaptive topologies. This iterative, guided synthesis process distinguishes GTD from single-step generative frameworks, enabling it to better navigate complex design trade-offs. We validated GTD across multiple benchmarks, and experiments show that this framework can generate highly task-adaptive, sparse, and efficient communication topologies, significantly outperforming existing methods in LLM agent collaboration.
FUSE: Measure-Theoretic Compact Fuzzy Set Representation for Taxonomy Expansion
Jun 10, 2025Abstract:Taxonomy Expansion, which models complex concepts and their relations, can be formulated as a set representation learning task. The generalization of set, fuzzy set, incorporates uncertainty and measures the information within a semantic concept, making it suitable for concept modeling. Existing works usually model sets as vectors or geometric objects such as boxes, which are not closed under set operations. In this work, we propose a sound and efficient formulation of set representation learning based on its volume approximation as a fuzzy set. The resulting embedding framework, Fuzzy Set Embedding (FUSE), satisfies all set operations and compactly approximates the underlying fuzzy set, hence preserving information while being efficient to learn, relying on minimum neural architecture. We empirically demonstrate the power of FUSE on the task of taxonomy expansion, where FUSE achieves remarkable improvements up to 23% compared with existing baselines. Our work marks the first attempt to understand and efficiently compute the embeddings of fuzzy sets.
FD-Bench: A Modular and Fair Benchmark for Data-driven Fluid Simulation
May 25, 2025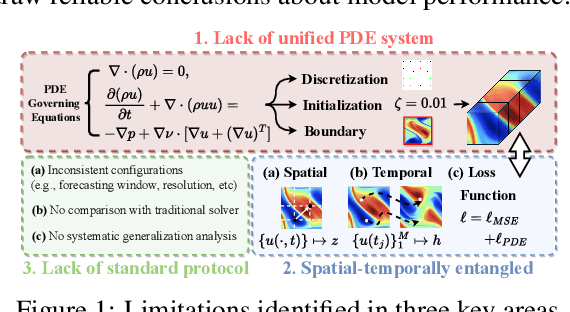
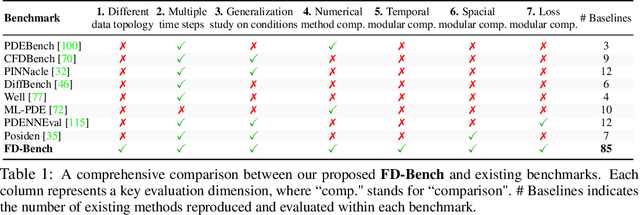
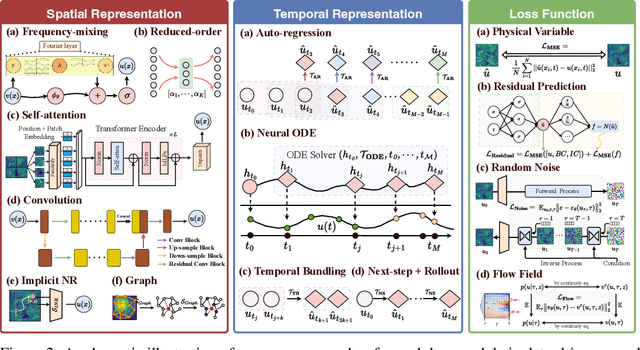
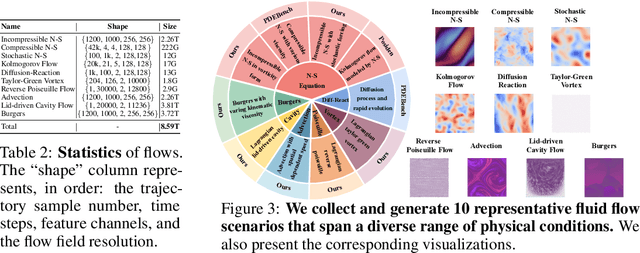
Abstract:Data-driven modeling of fluid dynamics has advanced rapidly with neural PDE solvers, yet a fair and strong benchmark remains fragmented due to the absence of unified PDE datasets and standardized evaluation protocols. Although architectural innovations are abundant, fair assessment is further impeded by the lack of clear disentanglement between spatial, temporal and loss modules. In this paper, we introduce FD-Bench, the first fair, modular, comprehensive and reproducible benchmark for data-driven fluid simulation. FD-Bench systematically evaluates 85 baseline models across 10 representative flow scenarios under a unified experimental setup. It provides four key contributions: (1) a modular design enabling fair comparisons across spatial, temporal, and loss function modules; (2) the first systematic framework for direct comparison with traditional numerical solvers; (3) fine-grained generalization analysis across resolutions, initial conditions, and temporal windows; and (4) a user-friendly, extensible codebase to support future research. Through rigorous empirical studies, FD-Bench establishes the most comprehensive leaderboard to date, resolving long-standing issues in reproducibility and comparability, and laying a foundation for robust evaluation of future data-driven fluid models. The code is open-sourced at https://anonymous.4open.science/r/FD-Bench-15BC.
LLM-DSE: Searching Accelerator Parameters with LLM Agents
May 18, 2025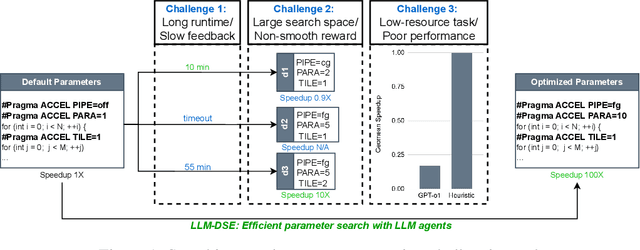
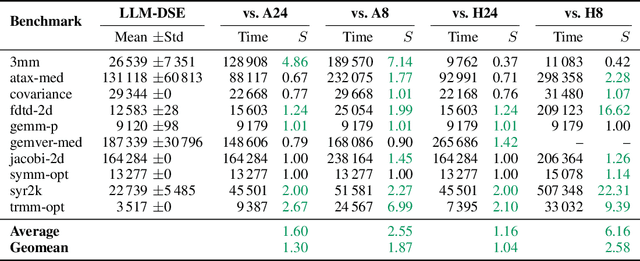
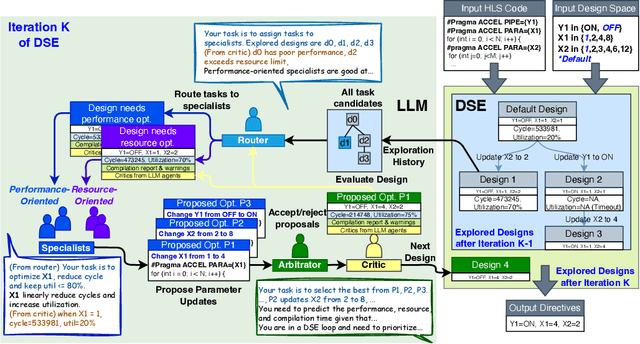
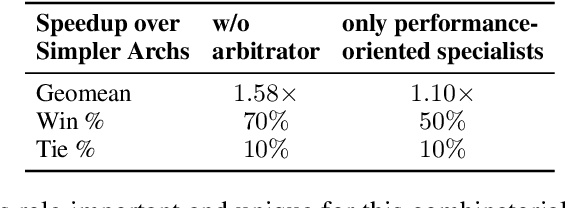
Abstract:Even though high-level synthesis (HLS) tools mitigate the challenges of programming domain-specific accelerators (DSAs) by raising the abstraction level, optimizing hardware directive parameters remains a significant hurdle. Existing heuristic and learning-based methods struggle with adaptability and sample efficiency.We present LLM-DSE, a multi-agent framework designed specifically for optimizing HLS directives. Combining LLM with design space exploration (DSE), our explorer coordinates four agents: Router, Specialists, Arbitrator, and Critic. These multi-agent components interact with various tools to accelerate the optimization process. LLM-DSE leverages essential domain knowledge to identify efficient parameter combinations while maintaining adaptability through verbal learning from online interactions. Evaluations on the HLSyn dataset demonstrate that LLM-DSE achieves substantial $2.55\times$ performance gains over state-of-the-art methods, uncovering novel designs while reducing runtime. Ablation studies validate the effectiveness and necessity of the proposed agent interactions. Our code is open-sourced here: https://github.com/Nozidoali/LLM-DSE.
LIFT: LLM-Based Pragma Insertion for HLS via GNN Supervised Fine-Tuning
Apr 29, 2025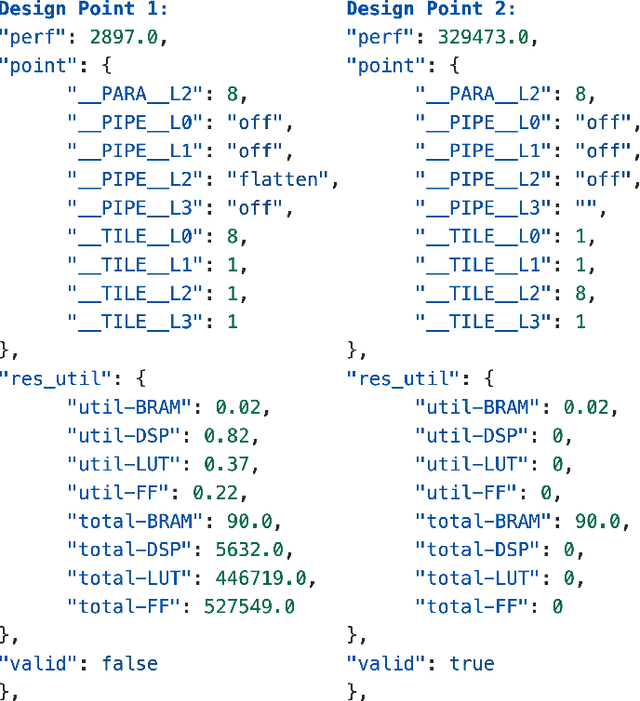
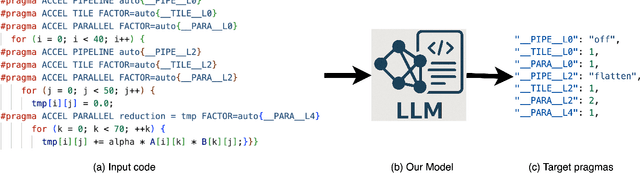
Abstract:FPGAs are increasingly adopted in datacenter environments for their reconfigurability and energy efficiency. High-Level Synthesis (HLS) tools have eased FPGA programming by raising the abstraction level from RTL to untimed C/C++, yet attaining high performance still demands expert knowledge and iterative manual insertion of optimization pragmas to modify the microarchitecture. To address this challenge, we propose LIFT, a large language model (LLM)-based coding assistant for HLS that automatically generates performance-critical pragmas given a C/C++ design. We fine-tune the LLM by tightly integrating and supervising the training process with a graph neural network (GNN), combining the sequential modeling capabilities of LLMs with the structural and semantic understanding of GNNs necessary for reasoning over code and its control/data dependencies. On average, LIFT produces designs that improve performance by 3.52x and 2.16x than prior state-of the art AutoDSE and HARP respectively, and 66x than GPT-4o.
Agree to Disagree? A Meta-Evaluation of LLM Misgendering
Apr 23, 2025Abstract:Numerous methods have been proposed to measure LLM misgendering, including probability-based evaluations (e.g., automatically with templatic sentences) and generation-based evaluations (e.g., with automatic heuristics or human validation). However, it has gone unexamined whether these evaluation methods have convergent validity, that is, whether their results align. Therefore, we conduct a systematic meta-evaluation of these methods across three existing datasets for LLM misgendering. We propose a method to transform each dataset to enable parallel probability- and generation-based evaluation. Then, by automatically evaluating a suite of 6 models from 3 families, we find that these methods can disagree with each other at the instance, dataset, and model levels, conflicting on 20.2% of evaluation instances. Finally, with a human evaluation of 2400 LLM generations, we show that misgendering behaviour is complex and goes far beyond pronouns, which automatic evaluations are not currently designed to capture, suggesting essential disagreement with human evaluations. Based on our findings, we provide recommendations for future evaluations of LLM misgendering. Our results are also more widely relevant, as they call into question broader methodological conventions in LLM evaluation, which often assume that different evaluation methods agree.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge