Yuanda Zhu
EHRAgent: Code Empowers Large Language Models for Complex Tabular Reasoning on Electronic Health Records
Jan 13, 2024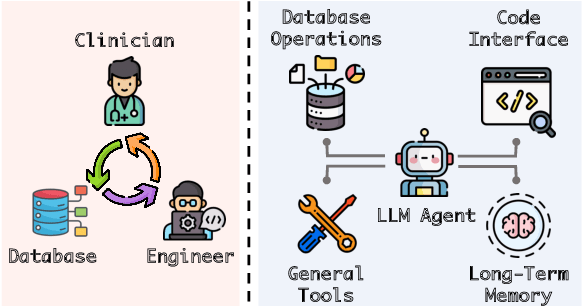
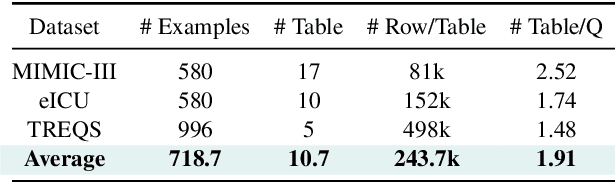
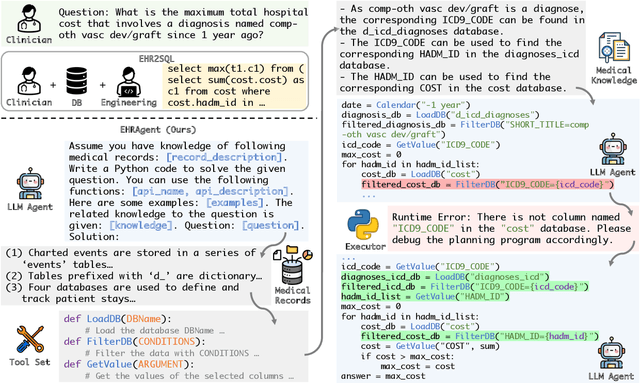
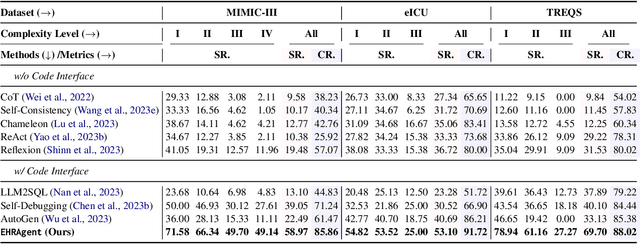
Abstract:Large language models (LLMs) have demonstrated exceptional capabilities in planning and tool utilization as autonomous agents, but few have been developed for medical problem-solving. We propose EHRAgent1, an LLM agent empowered with a code interface, to autonomously generate and execute code for complex clinical tasks within electronic health records (EHRs). First, we formulate an EHR question-answering task into a tool-use planning process, efficiently decomposing a complicated task into a sequence of manageable actions. By integrating interactive coding and execution feedback, EHRAgent learns from error messages and improves the originally generated code through iterations. Furthermore, we enhance the LLM agent by incorporating long-term memory, which allows EHRAgent to effectively select and build upon the most relevant successful cases from past experiences. Experiments on two real-world EHR datasets show that EHRAgent outperforms the strongest LLM agent baseline by 36.48% and 12.41%, respectively. EHRAgent leverages the emerging few-shot learning capabilities of LLMs, enabling autonomous code generation and execution to tackle complex clinical tasks with minimal demonstrations.
Explainable Artificial Intelligence Methods in Combating Pandemics: A Systematic Review
Dec 25, 2021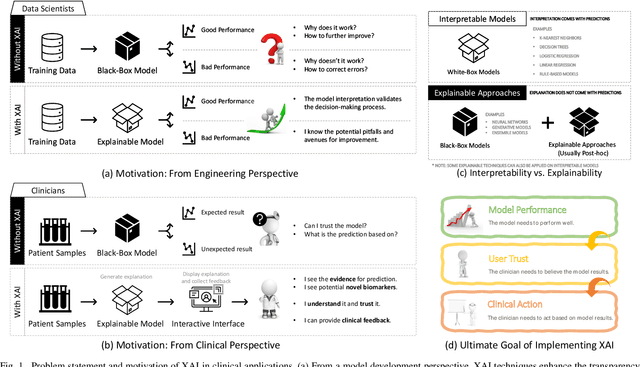
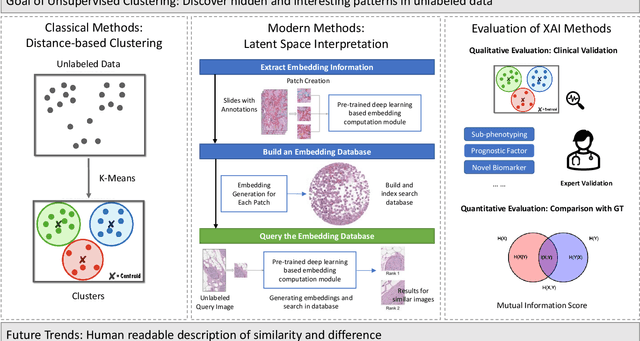
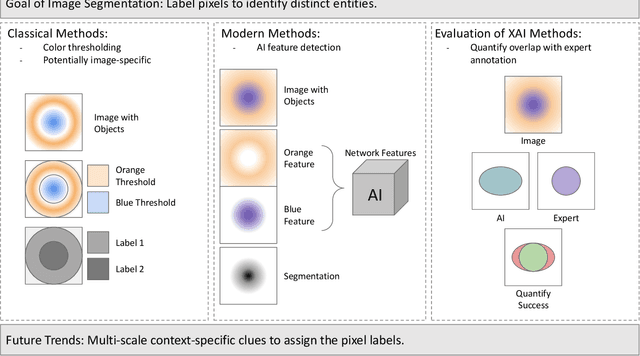
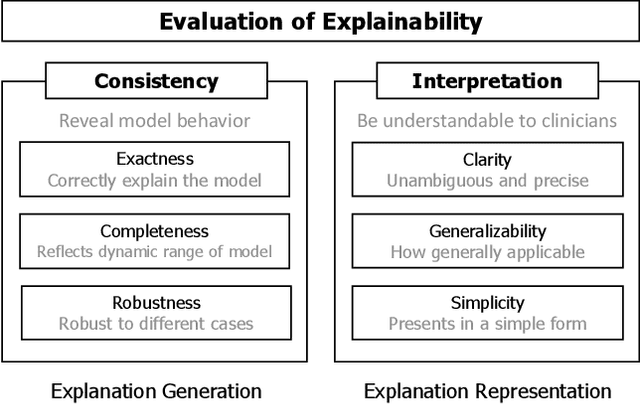
Abstract:Despite the myriad peer-reviewed papers demonstrating novel Artificial Intelligence (AI)-based solutions to COVID-19 challenges during the pandemic, few have made significant clinical impact. The impact of artificial intelligence during the COVID-19 pandemic was greatly limited by lack of model transparency. This systematic review examines the use of Explainable Artificial Intelligence (XAI) during the pandemic and how its use could overcome barriers to real-world success. We find that successful use of XAI can improve model performance, instill trust in the end-user, and provide the value needed to affect user decision-making. We introduce the reader to common XAI techniques, their utility, and specific examples of their application. Evaluation of XAI results is also discussed as an important step to maximize the value of AI-based clinical decision support systems. We illustrate the classical, modern, and potential future trends of XAI to elucidate the evolution of novel XAI techniques. Finally, we provide a checklist of suggestions during the experimental design process supported by recent publications. Common challenges during the implementation of AI solutions are also addressed with specific examples of potential solutions. We hope this review may serve as a guide to improve the clinical impact of future AI-based solutions.
Public Health Informatics: Proposing Causal Sequence of Death Using Neural Machine Translation
Sep 22, 2020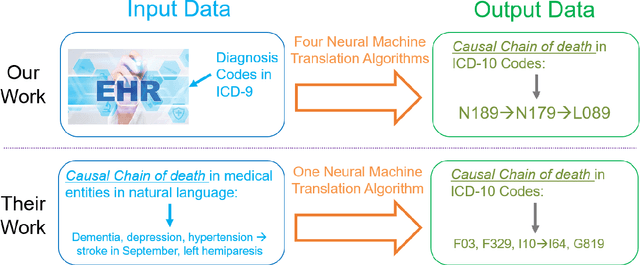
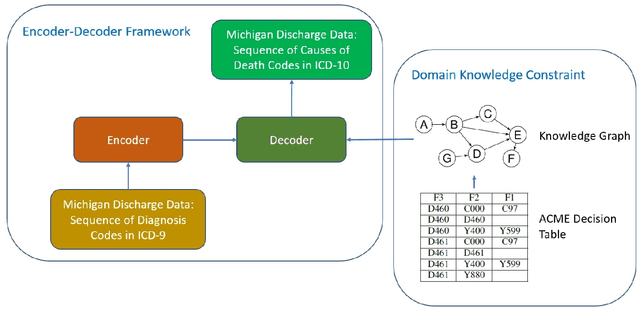
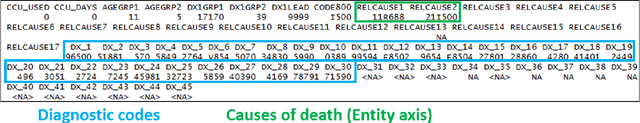
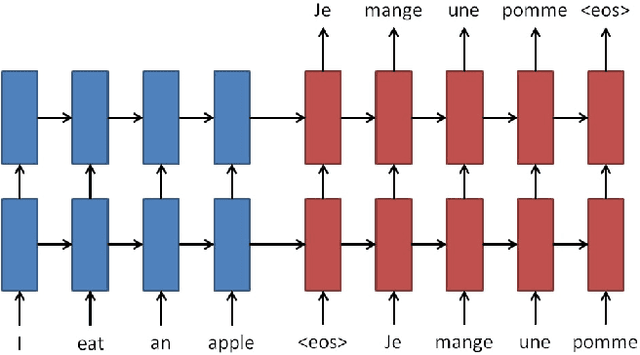
Abstract:Each year there are nearly 57 million deaths around the world, with over 2.7 million in the United States. Timely, accurate and complete death reporting is critical in public health, as institutions and government agencies rely on death reports to analyze vital statistics and to formulate responses to communicable diseases. Inaccurate death reporting may result in potential misdirection of public health policies. Determining the causes of death is, nevertheless, challenging even for experienced physicians. To facilitate physicians in accurately reporting causes of death, we present an advanced AI approach to determine a chronically ordered sequence of clinical conditions that lead to death, based on decedent's last hospital admission discharge record. The sequence of clinical codes on the death report is named as causal chain of death, coded in the tenth revision of International Statistical Classification of Diseases (ICD-10); the priority-ordered clinical conditions on the discharge record are coded in ICD-9. We identify three challenges in proposing the causal chain of death: two versions of coding system in clinical codes, medical domain knowledge conflict, and data interoperability. To overcome the first challenge in this sequence-to-sequence problem, we apply neural machine translation models to generate target sequence. We evaluate the quality of generated sequences with the BLEU (BiLingual Evaluation Understudy) score and achieve 16.44 out of 100. To address the second challenge, we incorporate expert-verified medical domain knowledge as constraint in generating output sequence to exclude infeasible causal chains. Lastly, we demonstrate the usability of our work in a Fast Healthcare Interoperability Resources (FHIR) interface to address the third challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge