Jonas Kemp
AfriMed-QA: A Pan-African, Multi-Specialty, Medical Question-Answering Benchmark Dataset
Nov 23, 2024

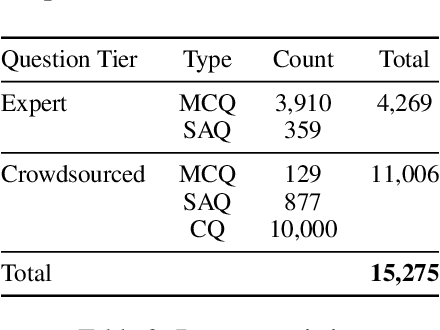

Abstract:Recent advancements in large language model(LLM) performance on medical multiple choice question (MCQ) benchmarks have stimulated interest from healthcare providers and patients globally. Particularly in low-and middle-income countries (LMICs) facing acute physician shortages and lack of specialists, LLMs offer a potentially scalable pathway to enhance healthcare access and reduce costs. However, their effectiveness in the Global South, especially across the African continent, remains to be established. In this work, we introduce AfriMed-QA, the first large scale Pan-African English multi-specialty medical Question-Answering (QA) dataset, 15,000 questions (open and closed-ended) sourced from over 60 medical schools across 16 countries, covering 32 medical specialties. We further evaluate 30 LLMs across multiple axes including correctness and demographic bias. Our findings show significant performance variation across specialties and geographies, MCQ performance clearly lags USMLE (MedQA). We find that biomedical LLMs underperform general models and smaller edge-friendly LLMs struggle to achieve a passing score. Interestingly, human evaluations show a consistent consumer preference for LLM answers and explanations when compared with clinician answers.
Capabilities of Gemini Models in Medicine
May 01, 2024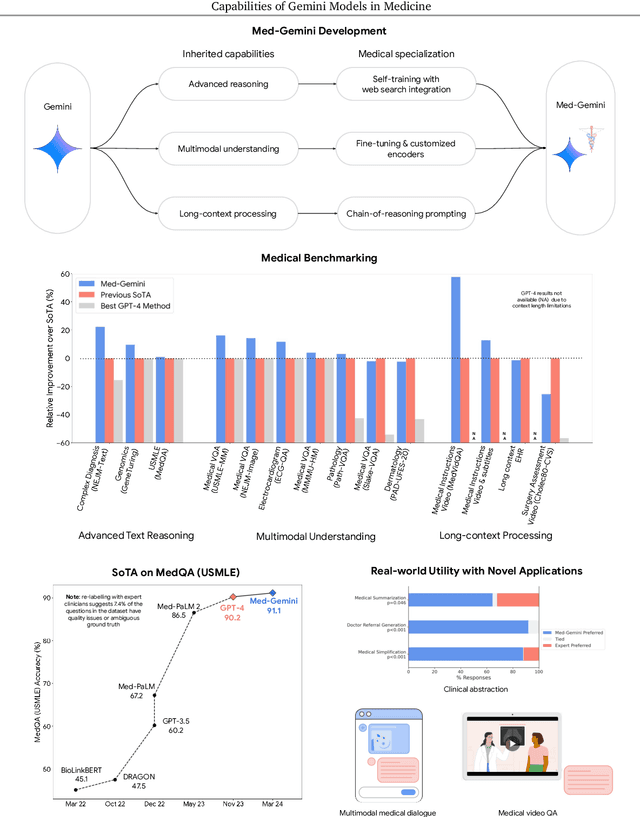
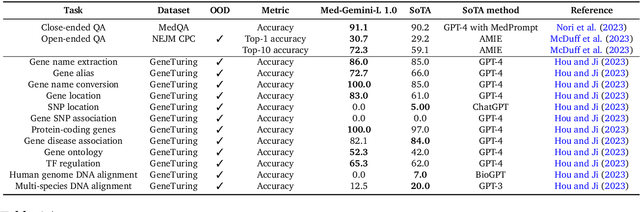
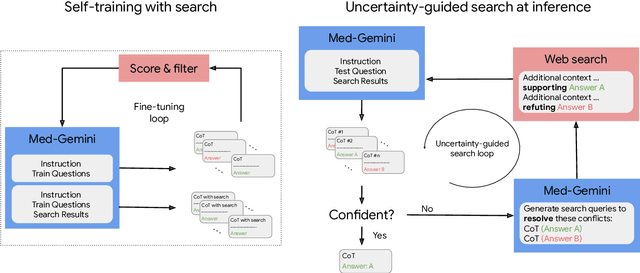
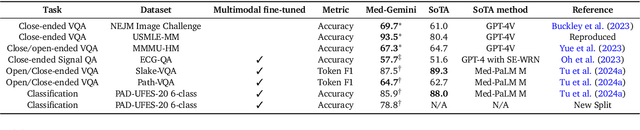
Abstract:Excellence in a wide variety of medical applications poses considerable challenges for AI, requiring advanced reasoning, access to up-to-date medical knowledge and understanding of complex multimodal data. Gemini models, with strong general capabilities in multimodal and long-context reasoning, offer exciting possibilities in medicine. Building on these core strengths of Gemini, we introduce Med-Gemini, a family of highly capable multimodal models that are specialized in medicine with the ability to seamlessly use web search, and that can be efficiently tailored to novel modalities using custom encoders. We evaluate Med-Gemini on 14 medical benchmarks, establishing new state-of-the-art (SoTA) performance on 10 of them, and surpass the GPT-4 model family on every benchmark where a direct comparison is viable, often by a wide margin. On the popular MedQA (USMLE) benchmark, our best-performing Med-Gemini model achieves SoTA performance of 91.1% accuracy, using a novel uncertainty-guided search strategy. On 7 multimodal benchmarks including NEJM Image Challenges and MMMU (health & medicine), Med-Gemini improves over GPT-4V by an average relative margin of 44.5%. We demonstrate the effectiveness of Med-Gemini's long-context capabilities through SoTA performance on a needle-in-a-haystack retrieval task from long de-identified health records and medical video question answering, surpassing prior bespoke methods using only in-context learning. Finally, Med-Gemini's performance suggests real-world utility by surpassing human experts on tasks such as medical text summarization, alongside demonstrations of promising potential for multimodal medical dialogue, medical research and education. Taken together, our results offer compelling evidence for Med-Gemini's potential, although further rigorous evaluation will be crucial before real-world deployment in this safety-critical domain.
Instability in clinical risk stratification models using deep learning
Nov 20, 2022



Abstract:While it has been well known in the ML community that deep learning models suffer from instability, the consequences for healthcare deployments are under characterised. We study the stability of different model architectures trained on electronic health records, using a set of outpatient prediction tasks as a case study. We show that repeated training runs of the same deep learning model on the same training data can result in significantly different outcomes at a patient level even though global performance metrics remain stable. We propose two stability metrics for measuring the effect of randomness of model training, as well as mitigation strategies for improving model stability.
Boosting the interpretability of clinical risk scores with intervention predictions
Jul 06, 2022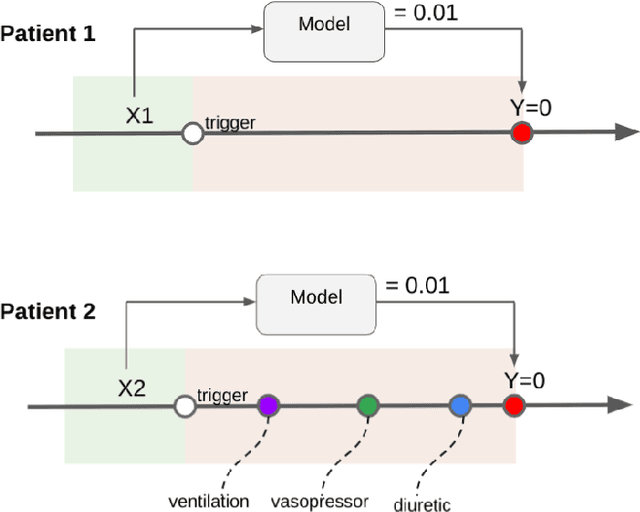
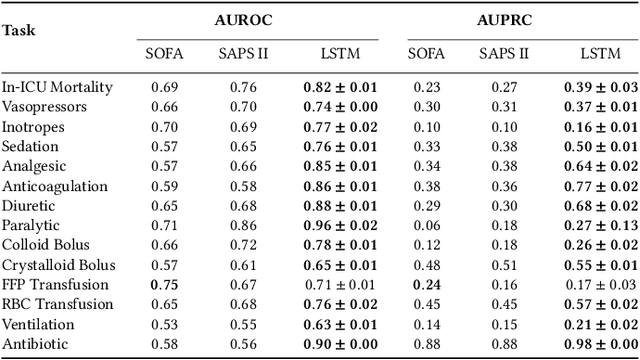
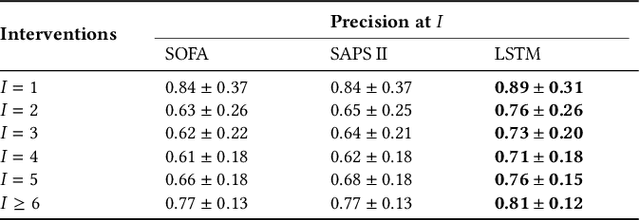
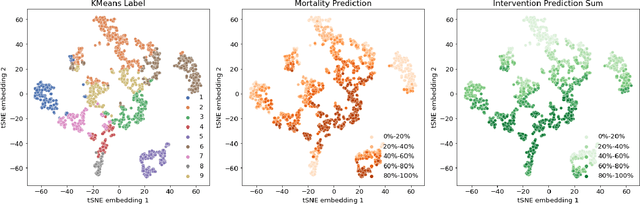
Abstract:Machine learning systems show significant promise for forecasting patient adverse events via risk scores. However, these risk scores implicitly encode assumptions about future interventions that the patient is likely to receive, based on the intervention policy present in the training data. Without this important context, predictions from such systems are less interpretable for clinicians. We propose a joint model of intervention policy and adverse event risk as a means to explicitly communicate the model's assumptions about future interventions. We develop such an intervention policy model on MIMIC-III, a real world de-identified ICU dataset, and discuss some use cases that highlight the utility of this approach. We show how combining typical risk scores, such as the likelihood of mortality, with future intervention probability scores leads to more interpretable clinical predictions.
Improved Patient Classification with Language Model Pretraining Over Clinical Notes
Oct 02, 2019



Abstract:Clinical notes in electronic health records contain highly heterogeneous writing styles, including non-standard terminology or abbreviations. Using these notes in predictive modeling has traditionally required preprocessing (e.g. taking frequent terms or topic modeling) that removes much of the richness of the source data. We propose a pretrained hierarchical recurrent neural network model that parses minimally processed clinical notes in an intuitive fashion, and show that it improves performance for multiple classification tasks on the Medical Information Mart for Intensive Care III (MIMIC-III) dataset, improving top-5 recall to 89.7% (increase of 4.8%) for primary diagnosis classification and AUPRC to 35.2% (increase of 2.1%) for multilabel diagnosis classification compared to models that treat the notes as an unordered collection of terms, using no pretraining. We also apply an attribution technique to several examples to identify the words and the nearby context that the model uses to make its prediction, and show the importance of the words' context.
Learning an Adaptive Learning Rate Schedule
Sep 20, 2019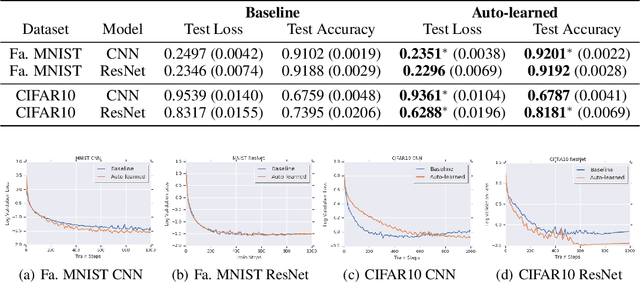
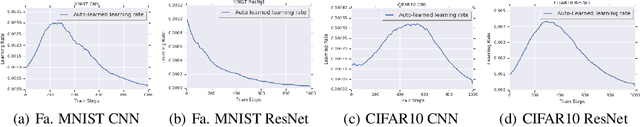
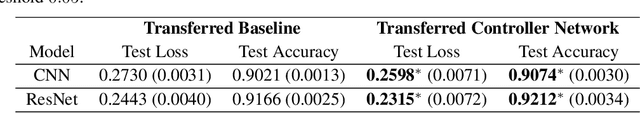
Abstract:The learning rate is one of the most important hyper-parameters for model training and generalization. However, current hand-designed parametric learning rate schedules offer limited flexibility and the predefined schedule may not match the training dynamics of high dimensional and non-convex optimization problems. In this paper, we propose a reinforcement learning based framework that can automatically learn an adaptive learning rate schedule by leveraging the information from past training histories. The learning rate dynamically changes based on the current training dynamics. To validate this framework, we conduct experiments with different neural network architectures on the Fashion MINIST and CIFAR10 datasets. Experimental results show that the auto-learned learning rate controller can achieve better test results. In addition, the trained controller network is generalizable -- able to be trained on one data set and transferred to new problems.
Analyzing the Role of Model Uncertainty for Electronic Health Records
Jun 10, 2019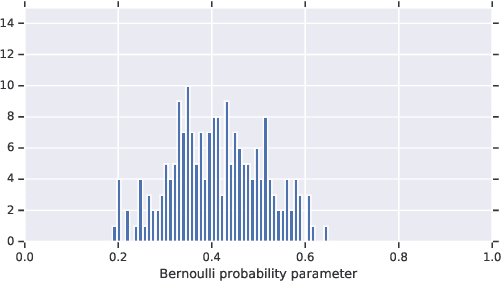
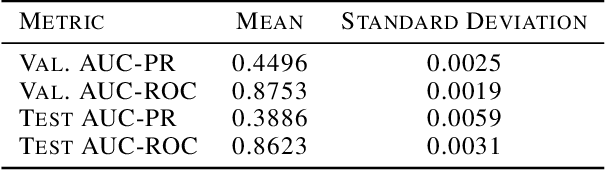
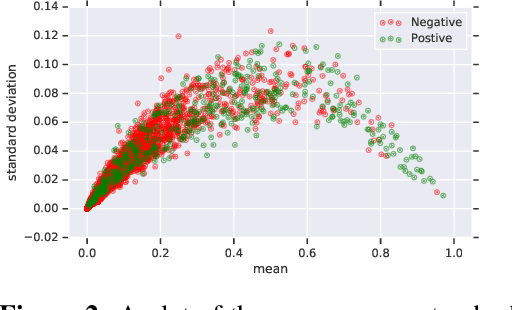
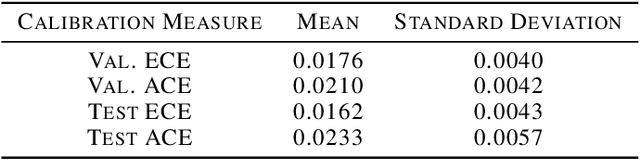
Abstract:In medicine, both ethical and monetary costs of incorrect predictions can be significant, and the complexity of the problems often necessitates increasingly complex models. Recent work has shown that changing just the random seed is enough for otherwise well-tuned deep neural networks to vary in their individual predicted probabilities. In light of this, we investigate the role of model uncertainty methods in the medical domain. Using RNN ensembles and various Bayesian RNNs, we show that population-level metrics, such as AUC-PR, AUC-ROC, log-likelihood, and calibration error, do not capture model uncertainty. Meanwhile, the presence of significant variability in patient-specific predictions and optimal decisions motivates the need for capturing model uncertainty. Understanding the uncertainty for individual patients is an area with clear clinical impact, such as determining when a model decision is likely to be brittle. We further show that RNNs with only Bayesian embeddings can be a more efficient way to capture model uncertainty compared to ensembles, and we analyze how model uncertainty is impacted across individual input features and patient subgroups.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge