Tassallah Abdullahi
UbuntuGuard: A Culturally-Grounded Policy Benchmark for Equitable AI Safety in African Languages
Jan 19, 2026Abstract:Current guardian models are predominantly Western-centric and optimized for high-resource languages, leaving low-resource African languages vulnerable to evolving harms, cross-lingual safety failures, and cultural misalignment. Moreover, most guardian models rely on rigid, predefined safety categories that fail to generalize across diverse linguistic and sociocultural contexts. Robust safety, therefore, requires flexible, runtime-enforceable policies and benchmarks that reflect local norms, harm scenarios, and cultural expectations. We introduce UbuntuGuard, the first African policy-based safety benchmark built from adversarial queries authored by 155 domain experts across sensitive fields, including healthcare. From these expert-crafted queries, we derive context-specific safety policies and reference responses that capture culturally grounded risk signals, enabling policy-aligned evaluation of guardian models. We evaluate 13 models, comprising six general-purpose LLMs and seven guardian models across three distinct variants: static, dynamic, and multilingual. Our findings reveal that existing English-centric benchmarks overestimate real-world multilingual safety, cross-lingual transfer provides partial but insufficient coverage, and dynamic models, while better equipped to leverage policies at inference time, still struggle to fully localize African-language contexts. These findings highlight the urgent need for multilingual, culturally grounded safety benchmarks to enable the development of reliable and equitable guardian models for low-resource languages. Our code can be found online.\footnote{Code repository available at https://github.com/hemhemoh/UbuntuGuard.
K-Paths: Reasoning over Graph Paths for Drug Repurposing and Drug Interaction Prediction
Feb 18, 2025Abstract:Drug discovery is a complex and time-intensive process that requires identifying and validating new therapeutic candidates. Computational approaches using large-scale biomedical knowledge graphs (KGs) offer a promising solution to accelerate this process. However, extracting meaningful insights from large-scale KGs remains challenging due to the complexity of graph traversal. Existing subgraph-based methods are tailored to graph neural networks (GNNs), making them incompatible with other models, such as large language models (LLMs). We introduce K-Paths, a retrieval framework that extracts structured, diverse, and biologically meaningful paths from KGs. Integrating these paths enables LLMs and GNNs to effectively predict unobserved drug-drug and drug-disease interactions. Unlike traditional path-ranking approaches, K-Paths retrieves and transforms paths into a structured format that LLMs can directly process, facilitating explainable reasoning. K-Paths employs a diversity-aware adaptation of Yen's algorithm to retrieve the K shortest loopless paths between entities in an interaction query, prioritizing biologically relevant and diverse relationships. Our experiments on benchmark datasets show that K-Paths improves the zero-shot performance of Llama 8.1B's F1-score by 12.45 points on drug repurposing and 13.42 points on interaction severity prediction. We also show that Llama 70B achieves F1-score gains of 6.18 and 8.46 points, respectively. K-Paths also improves the supervised training efficiency of EmerGNN, a state-of-the-art GNN, by reducing KG size by 90% while maintaining strong predictive performance. Beyond its scalability and efficiency, K-Paths uniquely bridges the gap between KGs and LLMs, providing explainable rationales for predicted interactions. These capabilities show that K-Paths is a valuable tool for efficient data-driven drug discovery.
Afrispeech-Dialog: A Benchmark Dataset for Spontaneous English Conversations in Healthcare and Beyond
Feb 06, 2025Abstract:Speech technologies are transforming interactions across various sectors, from healthcare to call centers and robots, yet their performance on African-accented conversations remains underexplored. We introduce Afrispeech-Dialog, a benchmark dataset of 50 simulated medical and non-medical African-accented English conversations, designed to evaluate automatic speech recognition (ASR) and related technologies. We assess state-of-the-art (SOTA) speaker diarization and ASR systems on long-form, accented speech, comparing their performance with native accents and discover a 10%+ performance degradation. Additionally, we explore medical conversation summarization capabilities of large language models (LLMs) to demonstrate the impact of ASR errors on downstream medical summaries, providing insights into the challenges and opportunities for speech technologies in the Global South. Our work highlights the need for more inclusive datasets to advance conversational AI in low-resource settings.
AfriMed-QA: A Pan-African, Multi-Specialty, Medical Question-Answering Benchmark Dataset
Nov 23, 2024

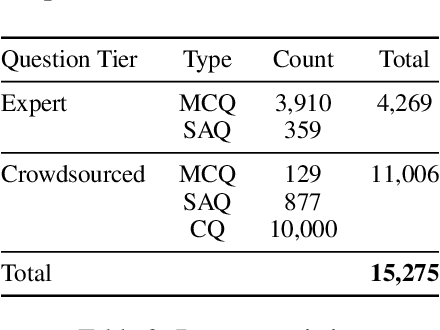

Abstract:Recent advancements in large language model(LLM) performance on medical multiple choice question (MCQ) benchmarks have stimulated interest from healthcare providers and patients globally. Particularly in low-and middle-income countries (LMICs) facing acute physician shortages and lack of specialists, LLMs offer a potentially scalable pathway to enhance healthcare access and reduce costs. However, their effectiveness in the Global South, especially across the African continent, remains to be established. In this work, we introduce AfriMed-QA, the first large scale Pan-African English multi-specialty medical Question-Answering (QA) dataset, 15,000 questions (open and closed-ended) sourced from over 60 medical schools across 16 countries, covering 32 medical specialties. We further evaluate 30 LLMs across multiple axes including correctness and demographic bias. Our findings show significant performance variation across specialties and geographies, MCQ performance clearly lags USMLE (MedQA). We find that biomedical LLMs underperform general models and smaller edge-friendly LLMs struggle to achieve a passing score. Interestingly, human evaluations show a consistent consumer preference for LLM answers and explanations when compared with clinician answers.
Retrieval Augmented Zero-Shot Text Classification
Jun 21, 2024



Abstract:Zero-shot text learning enables text classifiers to handle unseen classes efficiently, alleviating the need for task-specific training data. A simple approach often relies on comparing embeddings of query (text) to those of potential classes. However, the embeddings of a simple query sometimes lack rich contextual information, which hinders the classification performance. Traditionally, this has been addressed by improving the embedding model with expensive training. We introduce QZero, a novel training-free knowledge augmentation approach that reformulates queries by retrieving supporting categories from Wikipedia to improve zero-shot text classification performance. Our experiments across six diverse datasets demonstrate that QZero enhances performance for state-of-the-art static and contextual embedding models without the need for retraining. Notably, in News and medical topic classification tasks, QZero improves the performance of even the largest OpenAI embedding model by at least 5% and 3%, respectively. Acting as a knowledge amplifier, QZero enables small word embedding models to achieve performance levels comparable to those of larger contextual models, offering the potential for significant computational savings. Additionally, QZero offers meaningful insights that illuminate query context and verify topic relevance, aiding in understanding model predictions. Overall, QZero improves embedding-based zero-shot classifiers while maintaining their simplicity. This makes it particularly valuable for resource-constrained environments and domains with constantly evolving information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge