Tobi Olatunji
AfriSpeech-MultiBench: A Verticalized Multidomain Multicountry Benchmark Suite for African Accented English ASR
Nov 18, 2025Abstract:Recent advances in speech-enabled AI, including Google's NotebookLM and OpenAI's speech-to-speech API, are driving widespread interest in voice interfaces globally. Despite this momentum, there exists no publicly available application-specific model evaluation that caters to Africa's linguistic diversity. We present AfriSpeech-MultiBench, the first domain-specific evaluation suite for over 100 African English accents across 10+ countries and seven application domains: Finance, Legal, Medical, General dialogue, Call Center, Named Entities and Hallucination Robustness. We benchmark a diverse range of open, closed, unimodal ASR and multimodal LLM-based speech recognition systems using both spontaneous and non-spontaneous speech conversation drawn from various open African accented English speech datasets. Our empirical analysis reveals systematic variation: open-source ASR models excels in spontaneous speech contexts but degrades on noisy, non-native dialogue; multimodal LLMs are more accent-robust yet struggle with domain-specific named entities; proprietary models deliver high accuracy on clean speech but vary significantly by country and domain. Models fine-tuned on African English achieve competitive accuracy with lower latency, a practical advantage for deployment, hallucinations still remain a big problem for most SOTA models. By releasing this comprehensive benchmark, we empower practitioners and researchers to select voice technologies suited to African use-cases, fostering inclusive voice applications for underserved communities.
Afrispeech-Dialog: A Benchmark Dataset for Spontaneous English Conversations in Healthcare and Beyond
Feb 06, 2025Abstract:Speech technologies are transforming interactions across various sectors, from healthcare to call centers and robots, yet their performance on African-accented conversations remains underexplored. We introduce Afrispeech-Dialog, a benchmark dataset of 50 simulated medical and non-medical African-accented English conversations, designed to evaluate automatic speech recognition (ASR) and related technologies. We assess state-of-the-art (SOTA) speaker diarization and ASR systems on long-form, accented speech, comparing their performance with native accents and discover a 10%+ performance degradation. Additionally, we explore medical conversation summarization capabilities of large language models (LLMs) to demonstrate the impact of ASR errors on downstream medical summaries, providing insights into the challenges and opportunities for speech technologies in the Global South. Our work highlights the need for more inclusive datasets to advance conversational AI in low-resource settings.
The Multicultural Medical Assistant: Can LLMs Improve Medical ASR Errors Across Borders?
Jan 25, 2025Abstract:The global adoption of Large Language Models (LLMs) in healthcare shows promise to enhance clinical workflows and improve patient outcomes. However, Automatic Speech Recognition (ASR) errors in critical medical terms remain a significant challenge. These errors can compromise patient care and safety if not detected. This study investigates the prevalence and impact of ASR errors in medical transcription in Nigeria, the United Kingdom, and the United States. By evaluating raw and LLM-corrected transcriptions of accented English in these regions, we assess the potential and limitations of LLMs to address challenges related to accents and medical terminology in ASR. Our findings highlight significant disparities in ASR accuracy across regions and identify specific conditions under which LLM corrections are most effective.
AfriMed-QA: A Pan-African, Multi-Specialty, Medical Question-Answering Benchmark Dataset
Nov 23, 2024

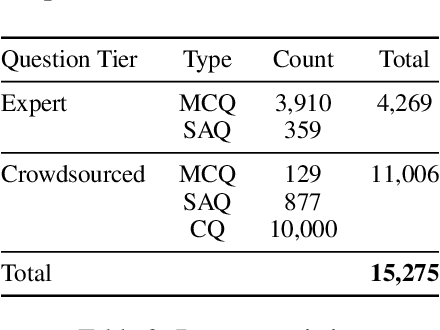

Abstract:Recent advancements in large language model(LLM) performance on medical multiple choice question (MCQ) benchmarks have stimulated interest from healthcare providers and patients globally. Particularly in low-and middle-income countries (LMICs) facing acute physician shortages and lack of specialists, LLMs offer a potentially scalable pathway to enhance healthcare access and reduce costs. However, their effectiveness in the Global South, especially across the African continent, remains to be established. In this work, we introduce AfriMed-QA, the first large scale Pan-African English multi-specialty medical Question-Answering (QA) dataset, 15,000 questions (open and closed-ended) sourced from over 60 medical schools across 16 countries, covering 32 medical specialties. We further evaluate 30 LLMs across multiple axes including correctness and demographic bias. Our findings show significant performance variation across specialties and geographies, MCQ performance clearly lags USMLE (MedQA). We find that biomedical LLMs underperform general models and smaller edge-friendly LLMs struggle to achieve a passing score. Interestingly, human evaluations show a consistent consumer preference for LLM answers and explanations when compared with clinician answers.
Performant ASR Models for Medical Entities in Accented Speech
Jun 18, 2024



Abstract:Recent strides in automatic speech recognition (ASR) have accelerated their application in the medical domain where their performance on accented medical named entities (NE) such as drug names, diagnoses, and lab results, is largely unknown. We rigorously evaluate multiple ASR models on a clinical English dataset of 93 African accents. Our analysis reveals that despite some models achieving low overall word error rates (WER), errors in clinical entities are higher, potentially posing substantial risks to patient safety. To empirically demonstrate this, we extract clinical entities from transcripts, develop a novel algorithm to align ASR predictions with these entities, and compute medical NE Recall, medical WER, and character error rate. Our results show that fine-tuning on accented clinical speech improves medical WER by a wide margin (25-34 % relative), improving their practical applicability in healthcare environments.
1000 African Voices: Advancing inclusive multi-speaker multi-accent speech synthesis
Jun 17, 2024Abstract:Recent advances in speech synthesis have enabled many useful applications like audio directions in Google Maps, screen readers, and automated content generation on platforms like TikTok. However, these systems are mostly dominated by voices sourced from data-rich geographies with personas representative of their source data. Although 3000 of the world's languages are domiciled in Africa, African voices and personas are under-represented in these systems. As speech synthesis becomes increasingly democratized, it is desirable to increase the representation of African English accents. We present Afro-TTS, the first pan-African accented English speech synthesis system able to generate speech in 86 African accents, with 1000 personas representing the rich phonological diversity across the continent for downstream application in Education, Public Health, and Automated Content Creation. Speaker interpolation retains naturalness and accentedness, enabling the creation of new voices.
AccentFold: A Journey through African Accents for Zero-Shot ASR Adaptation to Target Accents
Feb 05, 2024Abstract:Despite advancements in speech recognition, accented speech remains challenging. While previous approaches have focused on modeling techniques or creating accented speech datasets, gathering sufficient data for the multitude of accents, particularly in the African context, remains impractical due to their sheer diversity and associated budget constraints. To address these challenges, we propose AccentFold, a method that exploits spatial relationships between learned accent embeddings to improve downstream Automatic Speech Recognition (ASR). Our exploratory analysis of speech embeddings representing 100+ African accents reveals interesting spatial accent relationships highlighting geographic and genealogical similarities, capturing consistent phonological, and morphological regularities, all learned empirically from speech. Furthermore, we discover accent relationships previously uncharacterized by the Ethnologue. Through empirical evaluation, we demonstrate the effectiveness of AccentFold by showing that, for out-of-distribution (OOD) accents, sampling accent subsets for training based on AccentFold information outperforms strong baselines a relative WER improvement of 4.6%. AccentFold presents a promising approach for improving ASR performance on accented speech, particularly in the context of African accents, where data scarcity and budget constraints pose significant challenges. Our findings emphasize the potential of leveraging linguistic relationships to improve zero-shot ASR adaptation to target accents.
AfriSpeech-200: Pan-African Accented Speech Dataset for Clinical and General Domain ASR
Sep 30, 2023Abstract:Africa has a very low doctor-to-patient ratio. At very busy clinics, doctors could see 30+ patients per day -- a heavy patient burden compared with developed countries -- but productivity tools such as clinical automatic speech recognition (ASR) are lacking for these overworked clinicians. However, clinical ASR is mature, even ubiquitous, in developed nations, and clinician-reported performance of commercial clinical ASR systems is generally satisfactory. Furthermore, the recent performance of general domain ASR is approaching human accuracy. However, several gaps exist. Several publications have highlighted racial bias with speech-to-text algorithms and performance on minority accents lags significantly. To our knowledge, there is no publicly available research or benchmark on accented African clinical ASR, and speech data is non-existent for the majority of African accents. We release AfriSpeech, 200hrs of Pan-African English speech, 67,577 clips from 2,463 unique speakers across 120 indigenous accents from 13 countries for clinical and general domain ASR, a benchmark test set, with publicly available pre-trained models with SOTA performance on the AfriSpeech benchmark.
Adapting Pretrained ASR Models to Low-resource Clinical Speech using Epistemic Uncertainty-based Data Selection
Jun 03, 2023Abstract:While there has been significant progress in ASR, African-accented clinical ASR has been understudied due to a lack of training datasets. Building robust ASR systems in this domain requires large amounts of annotated or labeled data, for a wide variety of linguistically and morphologically rich accents, which are expensive to create. Our study aims to address this problem by reducing annotation expenses through informative uncertainty-based data selection. We show that incorporating epistemic uncertainty into our adaptation rounds outperforms several baseline results, established using state-of-the-art (SOTA) ASR models, while reducing the required amount of labeled data, and hence reducing annotation costs. Our approach also improves out-of-distribution generalization for very low-resource accents, demonstrating the viability of our approach for building generalizable ASR models in the context of accented African clinical ASR, where training datasets are predominantly scarce.
AfriNames: Most ASR models "butcher" African Names
Jun 02, 2023Abstract:Useful conversational agents must accurately capture named entities to minimize error for downstream tasks, for example, asking a voice assistant to play a track from a certain artist, initiating navigation to a specific location, or documenting a laboratory result for a patient. However, where named entities such as ``Ukachukwu`` (Igbo), ``Lakicia`` (Swahili), or ``Ingabire`` (Rwandan) are spoken, automatic speech recognition (ASR) models' performance degrades significantly, propagating errors to downstream systems. We model this problem as a distribution shift and demonstrate that such model bias can be mitigated through multilingual pre-training, intelligent data augmentation strategies to increase the representation of African-named entities, and fine-tuning multilingual ASR models on multiple African accents. The resulting fine-tuned models show an 81.5\% relative WER improvement compared with the baseline on samples with African-named entities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge