Ritambhara Singh
UbuntuGuard: A Culturally-Grounded Policy Benchmark for Equitable AI Safety in African Languages
Jan 19, 2026Abstract:Current guardian models are predominantly Western-centric and optimized for high-resource languages, leaving low-resource African languages vulnerable to evolving harms, cross-lingual safety failures, and cultural misalignment. Moreover, most guardian models rely on rigid, predefined safety categories that fail to generalize across diverse linguistic and sociocultural contexts. Robust safety, therefore, requires flexible, runtime-enforceable policies and benchmarks that reflect local norms, harm scenarios, and cultural expectations. We introduce UbuntuGuard, the first African policy-based safety benchmark built from adversarial queries authored by 155 domain experts across sensitive fields, including healthcare. From these expert-crafted queries, we derive context-specific safety policies and reference responses that capture culturally grounded risk signals, enabling policy-aligned evaluation of guardian models. We evaluate 13 models, comprising six general-purpose LLMs and seven guardian models across three distinct variants: static, dynamic, and multilingual. Our findings reveal that existing English-centric benchmarks overestimate real-world multilingual safety, cross-lingual transfer provides partial but insufficient coverage, and dynamic models, while better equipped to leverage policies at inference time, still struggle to fully localize African-language contexts. These findings highlight the urgent need for multilingual, culturally grounded safety benchmarks to enable the development of reliable and equitable guardian models for low-resource languages. Our code can be found online.\footnote{Code repository available at https://github.com/hemhemoh/UbuntuGuard.
Mechanisms of Prompt-Induced Hallucination in Vision-Language Models
Jan 08, 2026Abstract:Large vision-language models (VLMs) are highly capable, yet often hallucinate by favoring textual prompts over visual evidence. We study this failure mode in a controlled object-counting setting, where the prompt overstates the number of objects in the image (e.g., asking a model to describe four waterlilies when only three are present). At low object counts, models often correct the overestimation, but as the number of objects increases, they increasingly conform to the prompt regardless of the discrepancy. Through mechanistic analysis of three VLMs, we identify a small set of attention heads whose ablation substantially reduces prompt-induced hallucinations (PIH) by at least 40% without additional training. Across models, PIH-heads mediate prompt copying in model-specific ways. We characterize these differences and show that PIH ablation increases correction toward visual evidence. Our findings offer insights into the internal mechanisms driving prompt-induced hallucinations, revealing model-specific differences in how these behaviors are implemented.
Pixels Versus Priors: Controlling Knowledge Priors in Vision-Language Models through Visual Counterfacts
May 21, 2025Abstract:Multimodal Large Language Models (MLLMs) perform well on tasks such as visual question answering, but it remains unclear whether their reasoning relies more on memorized world knowledge or on the visual information present in the input image. To investigate this, we introduce Visual CounterFact, a new dataset of visually-realistic counterfactuals that put world knowledge priors (e.g, red strawberry) into direct conflict with visual input (e.g, blue strawberry). Using Visual CounterFact, we show that model predictions initially reflect memorized priors, but shift toward visual evidence in mid-to-late layers. This dynamic reveals a competition between the two modalities, with visual input ultimately overriding priors during evaluation. To control this behavior, we propose Pixels Versus Priors (PvP) steering vectors, a mechanism for controlling model outputs toward either world knowledge or visual input through activation-level interventions. On average, PvP successfully shifts 92.5% of color and 74.6% of size predictions from priors to counterfactuals. Together, these findings offer new tools for interpreting and controlling factual behavior in multimodal models.
Forgotten Polygons: Multimodal Large Language Models are Shape-Blind
Feb 21, 2025Abstract:Despite strong performance on vision-language tasks, Multimodal Large Language Models (MLLMs) struggle with mathematical problem-solving, with both open-source and state-of-the-art models falling short of human performance on visual-math benchmarks. To systematically examine visual-mathematical reasoning in MLLMs, we (1) evaluate their understanding of geometric primitives, (2) test multi-step reasoning, and (3) explore a potential solution to improve visual reasoning capabilities. Our findings reveal fundamental shortcomings in shape recognition, with top models achieving under 50% accuracy in identifying regular polygons. We analyze these failures through the lens of dual-process theory and show that MLLMs rely on System 1 (intuitive, memorized associations) rather than System 2 (deliberate reasoning). Consequently, MLLMs fail to count the sides of both familiar and novel shapes, suggesting they have neither learned the concept of sides nor effectively process visual inputs. Finally, we propose Visually Cued Chain-of-Thought (VC-CoT) prompting, which enhances multi-step mathematical reasoning by explicitly referencing visual annotations in diagrams, boosting GPT-4o's accuracy on an irregular polygon side-counting task from 7% to 93%. Our findings suggest that System 2 reasoning in MLLMs remains an open problem, and visually-guided prompting is essential for successfully engaging visual reasoning. Code available at: https://github.com/rsinghlab/Shape-Blind.
K-Paths: Reasoning over Graph Paths for Drug Repurposing and Drug Interaction Prediction
Feb 18, 2025Abstract:Drug discovery is a complex and time-intensive process that requires identifying and validating new therapeutic candidates. Computational approaches using large-scale biomedical knowledge graphs (KGs) offer a promising solution to accelerate this process. However, extracting meaningful insights from large-scale KGs remains challenging due to the complexity of graph traversal. Existing subgraph-based methods are tailored to graph neural networks (GNNs), making them incompatible with other models, such as large language models (LLMs). We introduce K-Paths, a retrieval framework that extracts structured, diverse, and biologically meaningful paths from KGs. Integrating these paths enables LLMs and GNNs to effectively predict unobserved drug-drug and drug-disease interactions. Unlike traditional path-ranking approaches, K-Paths retrieves and transforms paths into a structured format that LLMs can directly process, facilitating explainable reasoning. K-Paths employs a diversity-aware adaptation of Yen's algorithm to retrieve the K shortest loopless paths between entities in an interaction query, prioritizing biologically relevant and diverse relationships. Our experiments on benchmark datasets show that K-Paths improves the zero-shot performance of Llama 8.1B's F1-score by 12.45 points on drug repurposing and 13.42 points on interaction severity prediction. We also show that Llama 70B achieves F1-score gains of 6.18 and 8.46 points, respectively. K-Paths also improves the supervised training efficiency of EmerGNN, a state-of-the-art GNN, by reducing KG size by 90% while maintaining strong predictive performance. Beyond its scalability and efficiency, K-Paths uniquely bridges the gap between KGs and LLMs, providing explainable rationales for predicted interactions. These capabilities show that K-Paths is a valuable tool for efficient data-driven drug discovery.
BetaExplainer: A Probabilistic Method to Explain Graph Neural Networks
Dec 16, 2024Abstract:Graph neural networks (GNNs) are powerful tools for conducting inference on graph data but are often seen as "black boxes" due to difficulty in extracting meaningful subnetworks driving predictive performance. Many interpretable GNN methods exist, but they cannot quantify uncertainty in edge weights and suffer in predictive accuracy when applied to challenging graph structures. In this work, we proposed BetaExplainer which addresses these issues by using a sparsity-inducing prior to mask unimportant edges during model training. To evaluate our approach, we examine various simulated data sets with diverse real-world characteristics. Not only does this implementation provide a notion of edge importance uncertainty, it also improves upon evaluation metrics for challenging datasets compared to state-of-the art explainer methods.
What Do VLMs NOTICE? A Mechanistic Interpretability Pipeline for Noise-free Text-Image Corruption and Evaluation
Jun 24, 2024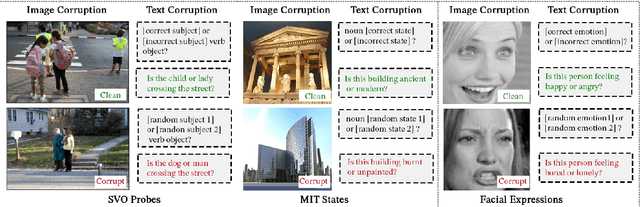

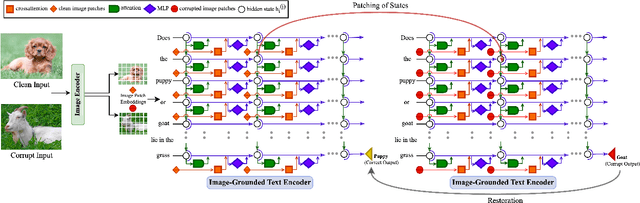
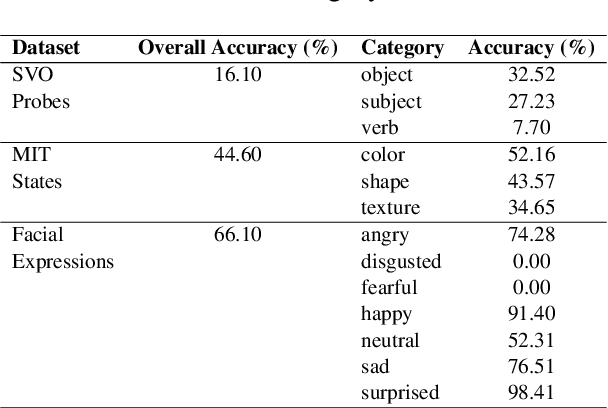
Abstract:Vision-Language Models (VLMs) have gained community-spanning prominence due to their ability to integrate visual and textual inputs to perform complex tasks. Despite their success, the internal decision-making processes of these models remain opaque, posing challenges in high-stakes applications. To address this, we introduce NOTICE, the first Noise-free Text-Image Corruption and Evaluation pipeline for mechanistic interpretability in VLMs. NOTICE incorporates a Semantic Minimal Pairs (SMP) framework for image corruption and Symmetric Token Replacement (STR) for text. This approach enables semantically meaningful causal mediation analysis for both modalities, providing a robust method for analyzing multimodal integration within models like BLIP. Our experiments on the SVO-Probes, MIT-States, and Facial Expression Recognition datasets reveal crucial insights into VLM decision-making, identifying the significant role of middle-layer cross-attention heads. Further, we uncover a set of ``universal cross-attention heads'' that consistently contribute across tasks and modalities, each performing distinct functions such as implicit image segmentation, object inhibition, and outlier inhibition. This work paves the way for more transparent and interpretable multimodal systems.
Retrieval Augmented Zero-Shot Text Classification
Jun 21, 2024



Abstract:Zero-shot text learning enables text classifiers to handle unseen classes efficiently, alleviating the need for task-specific training data. A simple approach often relies on comparing embeddings of query (text) to those of potential classes. However, the embeddings of a simple query sometimes lack rich contextual information, which hinders the classification performance. Traditionally, this has been addressed by improving the embedding model with expensive training. We introduce QZero, a novel training-free knowledge augmentation approach that reformulates queries by retrieving supporting categories from Wikipedia to improve zero-shot text classification performance. Our experiments across six diverse datasets demonstrate that QZero enhances performance for state-of-the-art static and contextual embedding models without the need for retraining. Notably, in News and medical topic classification tasks, QZero improves the performance of even the largest OpenAI embedding model by at least 5% and 3%, respectively. Acting as a knowledge amplifier, QZero enables small word embedding models to achieve performance levels comparable to those of larger contextual models, offering the potential for significant computational savings. Additionally, QZero offers meaningful insights that illuminate query context and verify topic relevance, aiding in understanding model predictions. Overall, QZero improves embedding-based zero-shot classifiers while maintaining their simplicity. This makes it particularly valuable for resource-constrained environments and domains with constantly evolving information.
On the Effect of Image Resolution on Semantic Segmentation
Feb 08, 2024Abstract:High-resolution semantic segmentation requires substantial computational resources. Traditional approaches in the field typically downscale the input images before processing and then upscale the low-resolution outputs back to their original dimensions. While this strategy effectively identifies broad regions, it often misses finer details. In this study, we demonstrate that a streamlined model capable of directly producing high-resolution segmentations can match the performance of more complex systems that generate lower-resolution results. By simplifying the network architecture, we enable the processing of images at their native resolution. Our approach leverages a bottom-up information propagation technique across various scales, which we have empirically shown to enhance segmentation accuracy. We have rigorously tested our method using leading-edge semantic segmentation datasets. Specifically, for the Cityscapes dataset, we further boost accuracy by applying the Noisy Student Training technique.
Revisiting invariances and introducing priors in Gromov-Wasserstein distances
Jul 19, 2023Abstract:Gromov-Wasserstein distance has found many applications in machine learning due to its ability to compare measures across metric spaces and its invariance to isometric transformations. However, in certain applications, this invariance property can be too flexible, thus undesirable. Moreover, the Gromov-Wasserstein distance solely considers pairwise sample similarities in input datasets, disregarding the raw feature representations. We propose a new optimal transport-based distance, called Augmented Gromov-Wasserstein, that allows for some control over the level of rigidity to transformations. It also incorporates feature alignments, enabling us to better leverage prior knowledge on the input data for improved performance. We present theoretical insights into the proposed metric. We then demonstrate its usefulness for single-cell multi-omic alignment tasks and a transfer learning scenario in machine learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge