Eric Loreaux
SDOH-NLI: a Dataset for Inferring Social Determinants of Health from Clinical Notes
Oct 27, 2023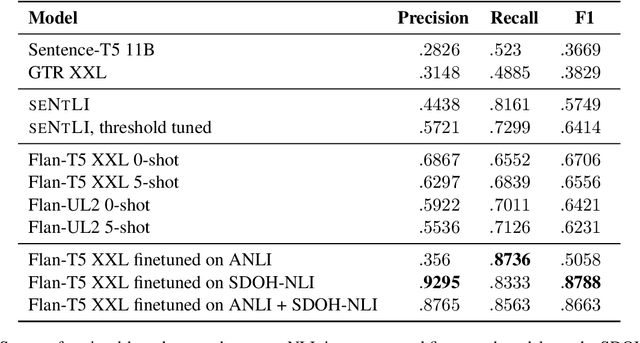
Abstract:Social and behavioral determinants of health (SDOH) play a significant role in shaping health outcomes, and extracting these determinants from clinical notes is a first step to help healthcare providers systematically identify opportunities to provide appropriate care and address disparities. Progress on using NLP methods for this task has been hindered by the lack of high-quality publicly available labeled data, largely due to the privacy and regulatory constraints on the use of real patients' information. This paper introduces a new dataset, SDOH-NLI, that is based on publicly available notes and which we release publicly. We formulate SDOH extraction as a natural language inference (NLI) task, and provide binary textual entailment labels obtained from human raters for a cross product of a set of social history snippets as premises and SDOH factors as hypotheses. Our dataset differs from standard NLI benchmarks in that our premises and hypotheses are obtained independently. We evaluate both "off-the-shelf" entailment models as well as models fine-tuned on our data, and highlight the ways in which our dataset appears more challenging than commonly used NLI datasets.
Boosting the interpretability of clinical risk scores with intervention predictions
Jul 06, 2022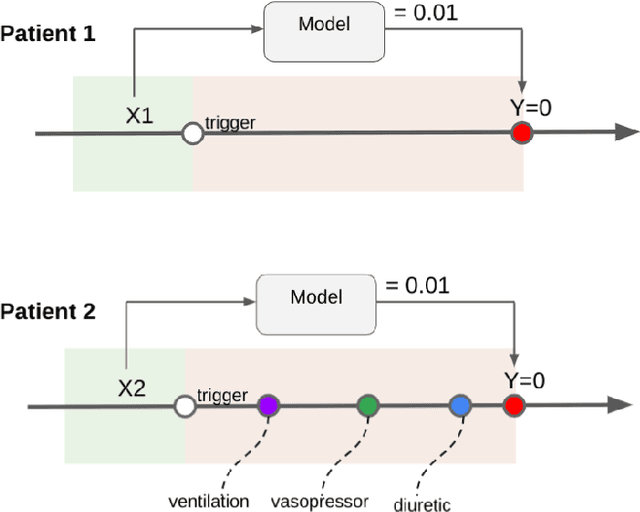
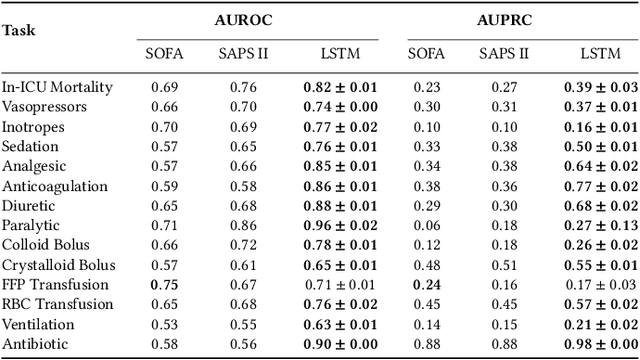
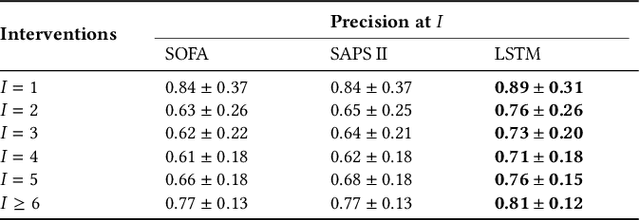
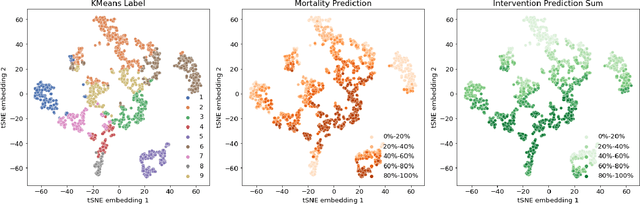
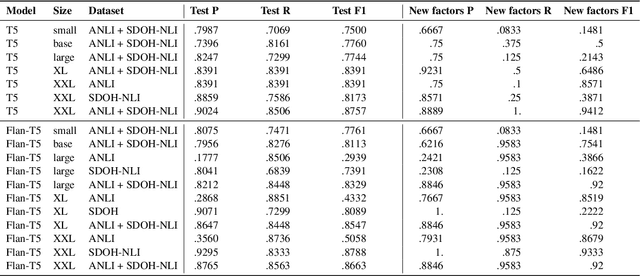
Abstract:Machine learning systems show significant promise for forecasting patient adverse events via risk scores. However, these risk scores implicitly encode assumptions about future interventions that the patient is likely to receive, based on the intervention policy present in the training data. Without this important context, predictions from such systems are less interpretable for clinicians. We propose a joint model of intervention policy and adverse event risk as a means to explicitly communicate the model's assumptions about future interventions. We develop such an intervention policy model on MIMIC-III, a real world de-identified ICU dataset, and discuss some use cases that highlight the utility of this approach. We show how combining typical risk scores, such as the likelihood of mortality, with future intervention probability scores leads to more interpretable clinical predictions.
Best of both worlds: local and global explanations with human-understandable concepts
Jun 16, 2021



Abstract:Interpretability techniques aim to provide the rationale behind a model's decision, typically by explaining either an individual prediction (local explanation, e.g. `why is this patient diagnosed with this condition') or a class of predictions (global explanation, e.g. `why are patients diagnosed with this condition in general'). While there are many methods focused on either one, few frameworks can provide both local and global explanations in a consistent manner. In this work, we combine two powerful existing techniques, one local (Integrated Gradients, IG) and one global (Testing with Concept Activation Vectors), to provide local, and global concept-based explanations. We first validate our idea using two synthetic datasets with a known ground truth, and further demonstrate with a benchmark natural image dataset. We test our method with various concepts, target classes, model architectures and IG baselines. We show that our method improves global explanations over TCAV when compared to ground truth, and provides useful insights. We hope our work provides a step towards building bridges between many existing local and global methods to get the best of both worlds.
Concept-based model explanations for Electronic Health Records
Dec 03, 2020



Abstract:Recurrent Neural Networks (RNNs) are often used for sequential modeling of adverse outcomes in electronic health records (EHRs) due to their ability to encode past clinical states. These deep, recurrent architectures have displayed increased performance compared to other modeling approaches in a number of tasks, fueling the interest in deploying deep models in clinical settings. One of the key elements in ensuring safe model deployment and building user trust is model explainability. Testing with Concept Activation Vectors (TCAV) has recently been introduced as a way of providing human-understandable explanations by comparing high-level concepts to the network's gradients. While the technique has shown promising results in real-world imaging applications, it has not been applied to structured temporal inputs. To enable an application of TCAV to sequential predictions in the EHR, we propose an extension of the method to time series data. We evaluate the proposed approach on an open EHR benchmark from the intensive care unit, as well as synthetic data where we are able to better isolate individual effects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge