Guoqing Liu
A Scientific Reasoning Model for Organic Synthesis Procedure Generation
Dec 15, 2025Abstract:Solving computer-aided synthesis planning is essential for enabling fully automated, robot-assisted synthesis workflows and improving the efficiency of drug discovery. A key challenge, however, is bridging the gap between computational route design and practical laboratory execution, particularly the accurate prediction of viable experimental procedures for each synthesis step. In this work, we present QFANG, a scientific reasoning language model capable of generating precise, structured experimental procedures directly from reaction equations, with explicit chain-of-thought reasoning. To develop QFANG, we curated a high-quality dataset comprising 905,990 chemical reactions paired with structured action sequences, extracted and processed from patent literature using large language models. We introduce a Chemistry-Guided Reasoning (CGR) framework that produces chain-of-thought data grounded in chemical knowledge at scale. The model subsequently undergoes supervised fine-tuning to elicit complex chemistry reasoning. Finally, we apply Reinforcement Learning from Verifiable Rewards (RLVR) to further enhance procedural accuracy. Experimental results demonstrate that QFANG outperforms advanced general-purpose reasoning models and nearest-neighbor retrieval baselines, measured by traditional NLP similarity metrics and a chemically aware evaluator using an LLM-as-a-judge. Moreover, QFANG generalizes to certain out-of-domain reaction classes and adapts to variations in laboratory conditions and user-specific constraints. We believe that QFANG's ability to generate high-quality synthesis procedures represents an important step toward bridging the gap between computational synthesis planning and fully automated laboratory synthesis.
Trust Region Preference Approximation: A simple and stable reinforcement learning algorithm for LLM reasoning
Apr 06, 2025
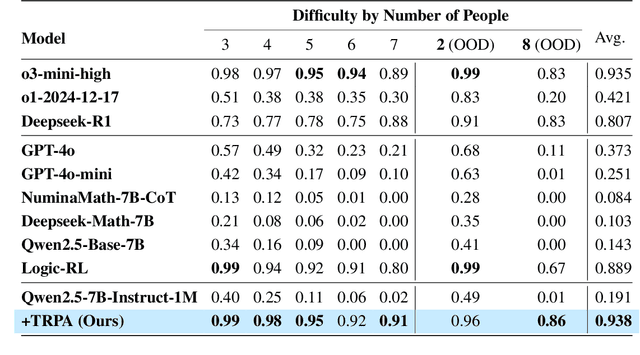
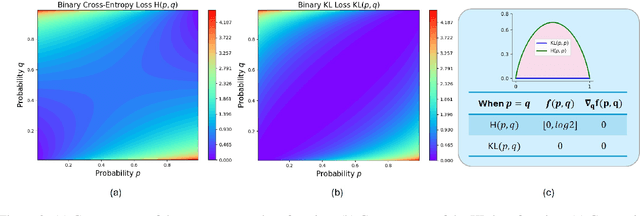
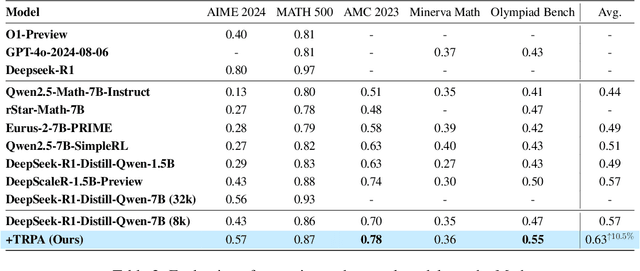
Abstract:Recently, Large Language Models (LLMs) have rapidly evolved, approaching Artificial General Intelligence (AGI) while benefiting from large-scale reinforcement learning to enhance Human Alignment (HA) and Reasoning. Recent reward-based optimization algorithms, such as Proximal Policy Optimization (PPO) and Group Relative Policy Optimization (GRPO) have achieved significant performance on reasoning tasks, whereas preference-based optimization algorithms such as Direct Preference Optimization (DPO) significantly improve the performance of LLMs on human alignment. However, despite the strong performance of reward-based optimization methods in alignment tasks , they remain vulnerable to reward hacking. Furthermore, preference-based algorithms (such as Online DPO) haven't yet matched the performance of reward-based optimization algorithms (like PPO) on reasoning tasks, making their exploration in this specific area still a worthwhile pursuit. Motivated by these challenges, we propose the Trust Region Preference Approximation (TRPA) algorithm, which integrates rule-based optimization with preference-based optimization for reasoning tasks. As a preference-based algorithm, TRPA naturally eliminates the reward hacking issue. TRPA constructs preference levels using predefined rules, forms corresponding preference pairs, and leverages a novel optimization algorithm for RL training with a theoretical monotonic improvement guarantee. Experimental results demonstrate that TRPA not only achieves competitive performance on reasoning tasks but also exhibits robust stability. The code of this paper are released and updating on https://github.com/XueruiSu/Trust-Region-Preference-Approximation.git.
TransDiffSBDD: Causality-Aware Multi-Modal Structure-Based Drug Design
Mar 26, 2025Abstract:Structure-based drug design (SBDD) is a critical task in drug discovery, requiring the generation of molecular information across two distinct modalities: discrete molecular graphs and continuous 3D coordinates. However, existing SBDD methods often overlook two key challenges: (1) the multi-modal nature of this task and (2) the causal relationship between these modalities, limiting their plausibility and performance. To address both challenges, we propose TransDiffSBDD, an integrated framework combining autoregressive transformers and diffusion models for SBDD. Specifically, the autoregressive transformer models discrete molecular information, while the diffusion model samples continuous distributions, effectively resolving the first challenge. To address the second challenge, we design a hybrid-modal sequence for protein-ligand complexes that explicitly respects the causality between modalities. Experiments on the CrossDocked2020 benchmark demonstrate that TransDiffSBDD outperforms existing baselines.
UniGenX: Unified Generation of Sequence and Structure with Autoregressive Diffusion
Mar 09, 2025Abstract:Unified generation of sequence and structure for scientific data (e.g., materials, molecules, proteins) is a critical task. Existing approaches primarily rely on either autoregressive sequence models or diffusion models, each offering distinct advantages and facing notable limitations. Autoregressive models, such as GPT, Llama, and Phi-4, have demonstrated remarkable success in natural language generation and have been extended to multimodal tasks (e.g., image, video, and audio) using advanced encoders like VQ-VAE to represent complex modalities as discrete sequences. However, their direct application to scientific domains is challenging due to the high precision requirements and the diverse nature of scientific data. On the other hand, diffusion models excel at generating high-dimensional scientific data, such as protein, molecule, and material structures, with remarkable accuracy. Yet, their inability to effectively model sequences limits their potential as general-purpose multimodal foundation models. To address these challenges, we propose UniGenX, a unified framework that combines autoregressive next-token prediction with conditional diffusion models. This integration leverages the strengths of autoregressive models to ease the training of conditional diffusion models, while diffusion-based generative heads enhance the precision of autoregressive predictions. We validate the effectiveness of UniGenX on material and small molecule generation tasks, achieving a significant leap in state-of-the-art performance for material crystal structure prediction and establishing new state-of-the-art results for small molecule structure prediction, de novo design, and conditional generation. Notably, UniGenX demonstrates significant improvements, especially in handling long sequences for complex structures, showcasing its efficacy as a versatile tool for scientific data generation.
NatureLM: Deciphering the Language of Nature for Scientific Discovery
Feb 11, 2025



Abstract:Foundation models have revolutionized natural language processing and artificial intelligence, significantly enhancing how machines comprehend and generate human languages. Inspired by the success of these foundation models, researchers have developed foundation models for individual scientific domains, including small molecules, materials, proteins, DNA, and RNA. However, these models are typically trained in isolation, lacking the ability to integrate across different scientific domains. Recognizing that entities within these domains can all be represented as sequences, which together form the "language of nature", we introduce Nature Language Model (briefly, NatureLM), a sequence-based science foundation model designed for scientific discovery. Pre-trained with data from multiple scientific domains, NatureLM offers a unified, versatile model that enables various applications including: (i) generating and optimizing small molecules, proteins, RNA, and materials using text instructions; (ii) cross-domain generation/design, such as protein-to-molecule and protein-to-RNA generation; and (iii) achieving state-of-the-art performance in tasks like SMILES-to-IUPAC translation and retrosynthesis on USPTO-50k. NatureLM offers a promising generalist approach for various scientific tasks, including drug discovery (hit generation/optimization, ADMET optimization, synthesis), novel material design, and the development of therapeutic proteins or nucleotides. We have developed NatureLM models in different sizes (1 billion, 8 billion, and 46.7 billion parameters) and observed a clear improvement in performance as the model size increases.
ARIES: Stimulating Self-Refinement of Large Language Models by Iterative Preference Optimization
Feb 08, 2025Abstract:A truly intelligent Large Language Model (LLM) should be capable of correcting errors in its responses through external interactions. However, even the most advanced models often face challenges in improving their outputs. In this paper, we explore how to cultivate LLMs with the self-refinement capability through iterative preference training, and how this ability can be leveraged to improve model performance during inference. To this end, we introduce a novel post-training and inference framework, called ARIES: Adaptive Refinement and Iterative Enhancement Structure. This method iteratively performs preference training and self-refinement-based data collection. During training, ARIES strengthen the model's direct question-answering capability while simultaneously unlocking its self-refinement potential. During inference, ARIES harnesses this self-refinement capability to generate a series of progressively refined responses, which are then filtered using either the Reward Model Scoring or a simple yet effective Rule-Based Selection mechanism, specifically tailored to our approach, to construct a dataset for the next round of preference training. Experimental results demonstrate the remarkable performance of ARIES. When applied to the Llama-3.1-8B model and under the self-refinement setting, ARIES surpasses powerful models such as GPT-4o, achieving 62.3% length-controlled (LC) and a 63.3% raw win rates on AlpacaEval 2, outperforming Iterative DPO by 27.8% and 35.5% respectively, as well as a 50.3% win rate on Arena-Hard, surpassing Iterative DPO by 26.6%. Furthermore, ARIES consistently enhances performance on mathematical reasoning tasks like GSM8K and MATH.
3DMolFormer: A Dual-channel Framework for Structure-based Drug Discovery
Feb 07, 2025



Abstract:Structure-based drug discovery, encompassing the tasks of protein-ligand docking and pocket-aware 3D drug design, represents a core challenge in drug discovery. However, no existing work can deal with both tasks to effectively leverage the duality between them, and current methods for each task are hindered by challenges in modeling 3D information and the limitations of available data. To address these issues, we propose 3DMolFormer, a unified dual-channel transformer-based framework applicable to both docking and 3D drug design tasks, which exploits their duality by utilizing docking functionalities within the drug design process. Specifically, we represent 3D pocket-ligand complexes using parallel sequences of discrete tokens and continuous numbers, and we design a corresponding dual-channel transformer model to handle this format, thereby overcoming the challenges of 3D information modeling. Additionally, we alleviate data limitations through large-scale pre-training on a mixed dataset, followed by supervised and reinforcement learning fine-tuning techniques respectively tailored for the two tasks. Experimental results demonstrate that 3DMolFormer outperforms previous approaches in both protein-ligand docking and pocket-aware 3D drug design, highlighting its promising application in structure-based drug discovery. The code is available at: https://github.com/HXYfighter/3DMolFormer .
Chimera: Accurate retrosynthesis prediction by ensembling models with diverse inductive biases
Dec 06, 2024



Abstract:Planning and conducting chemical syntheses remains a major bottleneck in the discovery of functional small molecules, and prevents fully leveraging generative AI for molecular inverse design. While early work has shown that ML-based retrosynthesis models can predict reasonable routes, their low accuracy for less frequent, yet important reactions has been pointed out. As multi-step search algorithms are limited to reactions suggested by the underlying model, the applicability of those tools is inherently constrained by the accuracy of retrosynthesis prediction. Inspired by how chemists use different strategies to ideate reactions, we propose Chimera: a framework for building highly accurate reaction models that combine predictions from diverse sources with complementary inductive biases using a learning-based ensembling strategy. We instantiate the framework with two newly developed models, which already by themselves achieve state of the art in their categories. Through experiments across several orders of magnitude in data scale and time-splits, we show Chimera outperforms all major models by a large margin, owing both to the good individual performance of its constituents, but also to the scalability of our ensembling strategy. Moreover, we find that PhD-level organic chemists prefer predictions from Chimera over baselines in terms of quality. Finally, we transfer the largest-scale checkpoint to an internal dataset from a major pharmaceutical company, showing robust generalization under distribution shift. With the new dimension that our framework unlocks, we anticipate further acceleration in the development of even more accurate models.
Theoretically Guaranteed Distribution Adaptable Learning
Nov 05, 2024Abstract:In many open environment applications, data are collected in the form of a stream, which exhibits an evolving distribution over time. How to design algorithms to track these evolving data distributions with provable guarantees, particularly in terms of the generalization ability, remains a formidable challenge. To handle this crucial but rarely studied problem and take a further step toward robust artificial intelligence, we propose a novel framework called Distribution Adaptable Learning (DAL). It enables the model to effectively track the evolving data distributions. By Encoding Feature Marginal Distribution Information (EFMDI), we broke the limitations of optimal transport to characterize the environmental changes and enable model reuse across diverse data distributions. It can enhance the reusable and evolvable properties of DAL in accommodating evolving distributions. Furthermore, to obtain the model interpretability, we not only analyze the generalization error bound of the local step in the evolution process, but also investigate the generalization error bound associated with the entire classifier trajectory of the evolution based on the Fisher-Rao distance. For demonstration, we also present two special cases within the framework, together with their optimizations and convergence analyses. Experimental results over both synthetic and real-world data distribution evolving tasks validate the effectiveness and practical utility of the proposed framework.
EPContrast: Effective Point-level Contrastive Learning for Large-scale Point Cloud Understanding
Oct 22, 2024



Abstract:The acquisition of inductive bias through point-level contrastive learning holds paramount significance in point cloud pre-training. However, the square growth in computational requirements with the scale of the point cloud poses a substantial impediment to the practical deployment and execution. To address this challenge, this paper proposes an Effective Point-level Contrastive Learning method for large-scale point cloud understanding dubbed \textbf{EPContrast}, which consists of AGContrast and ChannelContrast. In practice, AGContrast constructs positive and negative pairs based on asymmetric granularity embedding, while ChannelContrast imposes contrastive supervision between channel feature maps. EPContrast offers point-level contrastive loss while concurrently mitigating the computational resource burden. The efficacy of EPContrast is substantiated through comprehensive validation on S3DIS and ScanNetV2, encompassing tasks such as semantic segmentation, instance segmentation, and object detection. In addition, rich ablation experiments demonstrate remarkable bias induction capabilities under label-efficient and one-epoch training settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge