Chuan Cao
RaCalNet: Radar Calibration Network for Sparse-Supervised Metric Depth Estimation
Jun 18, 2025Abstract:Dense metric depth estimation using millimeter-wave radar typically requires dense LiDAR supervision, generated via multi-frame projection and interpolation, to guide the learning of accurate depth from sparse radar measurements and RGB images. However, this paradigm is both costly and data-intensive. To address this, we propose RaCalNet, a novel framework that eliminates the need for dense supervision by using sparse LiDAR to supervise the learning of refined radar measurements, resulting in a supervision density of merely around 1% compared to dense-supervised methods. Unlike previous approaches that associate radar points with broad image regions and rely heavily on dense labels, RaCalNet first recalibrates and refines sparse radar points to construct accurate depth priors. These priors then serve as reliable anchors to guide monocular depth prediction, enabling metric-scale estimation without resorting to dense supervision. This design improves structural consistency and preserves fine details. Despite relying solely on sparse supervision, RaCalNet surpasses state-of-the-art dense-supervised methods, producing depth maps with clear object contours and fine-grained textures. Extensive experiments on the ZJU-4DRadarCam dataset and real-world deployment scenarios demonstrate its effectiveness, reducing RMSE by 35.30% and 34.89%, respectively.
SALT: A Flexible Semi-Automatic Labeling Tool for General LiDAR Point Clouds with Cross-Scene Adaptability and 4D Consistency
Mar 31, 2025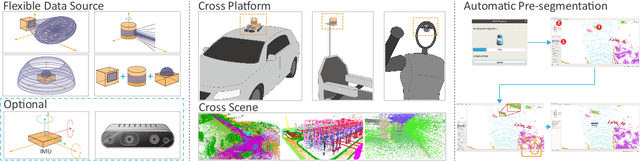
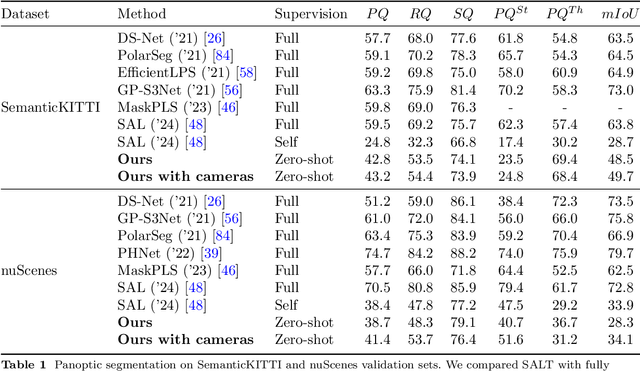
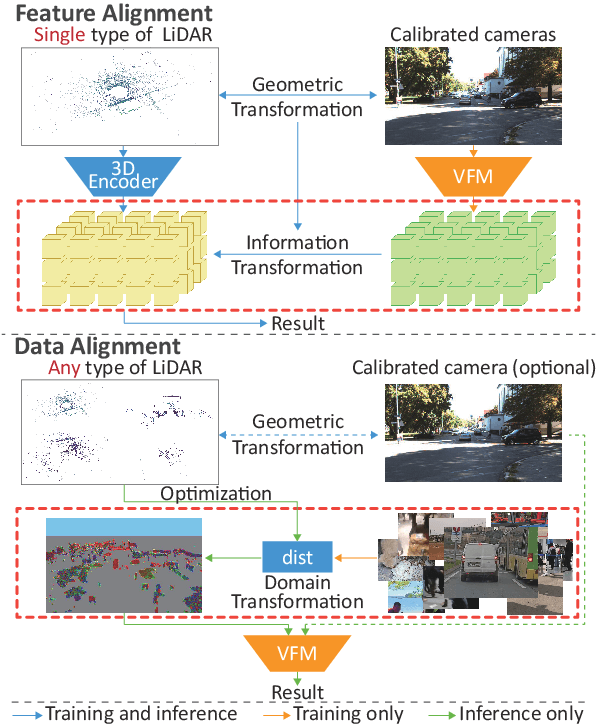
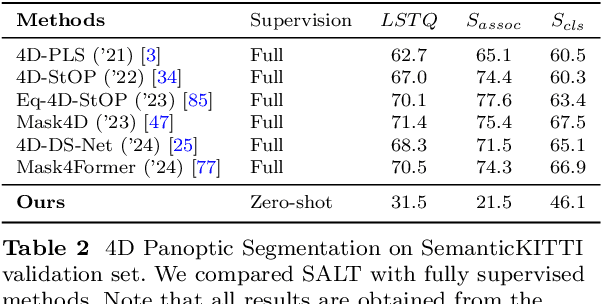
Abstract:We propose a flexible Semi-Automatic Labeling Tool (SALT) for general LiDAR point clouds with cross-scene adaptability and 4D consistency. Unlike recent approaches that rely on camera distillation, SALT operates directly on raw LiDAR data, automatically generating pre-segmentation results. To achieve this, we propose a novel zero-shot learning paradigm, termed data alignment, which transforms LiDAR data into pseudo-images by aligning with the training distribution of vision foundation models. Additionally, we design a 4D-consistent prompting strategy and 4D non-maximum suppression module to enhance SAM2, ensuring high-quality, temporally consistent presegmentation. SALT surpasses the latest zero-shot methods by 18.4% PQ on SemanticKITTI and achieves nearly 40-50% of human annotator performance on our newly collected low-resolution LiDAR data and on combined data from three LiDAR types, significantly boosting annotation efficiency. We anticipate that SALT's open-sourcing will catalyze substantial expansion of current LiDAR datasets and lay the groundwork for the future development of LiDAR foundation models. Code is available at https://github.com/Cavendish518/SALT.
Regulatory DNA sequence Design with Reinforcement Learning
Mar 11, 2025Abstract:Cis-regulatory elements (CREs), such as promoters and enhancers, are relatively short DNA sequences that directly regulate gene expression. The fitness of CREs, measured by their ability to modulate gene expression, highly depends on the nucleotide sequences, especially specific motifs known as transcription factor binding sites (TFBSs). Designing high-fitness CREs is crucial for therapeutic and bioengineering applications. Current CRE design methods are limited by two major drawbacks: (1) they typically rely on iterative optimization strategies that modify existing sequences and are prone to local optima, and (2) they lack the guidance of biological prior knowledge in sequence optimization. In this paper, we address these limitations by proposing a generative approach that leverages reinforcement learning (RL) to fine-tune a pre-trained autoregressive (AR) model. Our method incorporates data-driven biological priors by deriving computational inference-based rewards that simulate the addition of activator TFBSs and removal of repressor TFBSs, which are then integrated into the RL process. We evaluate our method on promoter design tasks in two yeast media conditions and enhancer design tasks for three human cell types, demonstrating its ability to generate high-fitness CREs while maintaining sequence diversity. The code is available at https://github.com/yangzhao1230/TACO.
NatureLM: Deciphering the Language of Nature for Scientific Discovery
Feb 11, 2025



Abstract:Foundation models have revolutionized natural language processing and artificial intelligence, significantly enhancing how machines comprehend and generate human languages. Inspired by the success of these foundation models, researchers have developed foundation models for individual scientific domains, including small molecules, materials, proteins, DNA, and RNA. However, these models are typically trained in isolation, lacking the ability to integrate across different scientific domains. Recognizing that entities within these domains can all be represented as sequences, which together form the "language of nature", we introduce Nature Language Model (briefly, NatureLM), a sequence-based science foundation model designed for scientific discovery. Pre-trained with data from multiple scientific domains, NatureLM offers a unified, versatile model that enables various applications including: (i) generating and optimizing small molecules, proteins, RNA, and materials using text instructions; (ii) cross-domain generation/design, such as protein-to-molecule and protein-to-RNA generation; and (iii) achieving state-of-the-art performance in tasks like SMILES-to-IUPAC translation and retrosynthesis on USPTO-50k. NatureLM offers a promising generalist approach for various scientific tasks, including drug discovery (hit generation/optimization, ADMET optimization, synthesis), novel material design, and the development of therapeutic proteins or nucleotides. We have developed NatureLM models in different sizes (1 billion, 8 billion, and 46.7 billion parameters) and observed a clear improvement in performance as the model size increases.
SFPNet: Sparse Focal Point Network for Semantic Segmentation on General LiDAR Point Clouds
Jul 16, 2024



Abstract:Although LiDAR semantic segmentation advances rapidly, state-of-the-art methods often incorporate specifically designed inductive bias derived from benchmarks originating from mechanical spinning LiDAR. This can limit model generalizability to other kinds of LiDAR technologies and make hyperparameter tuning more complex. To tackle these issues, we propose a generalized framework to accommodate various types of LiDAR prevalent in the market by replacing window-attention with our sparse focal point modulation. Our SFPNet is capable of extracting multi-level contexts and dynamically aggregating them using a gate mechanism. By implementing a channel-wise information query, features that incorporate both local and global contexts are encoded. We also introduce a novel large-scale hybrid-solid LiDAR semantic segmentation dataset for robotic applications. SFPNet demonstrates competitive performance on conventional benchmarks derived from mechanical spinning LiDAR, while achieving state-of-the-art results on benchmark derived from solid-state LiDAR. Additionally, it outperforms existing methods on our novel dataset sourced from hybrid-solid LiDAR. Code and dataset are available at https://github.com/Cavendish518/SFPNet and https://www.semanticindustry.top.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge