Krzysztof Maziarz
NatureLM: Deciphering the Language of Nature for Scientific Discovery
Feb 11, 2025



Abstract:Foundation models have revolutionized natural language processing and artificial intelligence, significantly enhancing how machines comprehend and generate human languages. Inspired by the success of these foundation models, researchers have developed foundation models for individual scientific domains, including small molecules, materials, proteins, DNA, and RNA. However, these models are typically trained in isolation, lacking the ability to integrate across different scientific domains. Recognizing that entities within these domains can all be represented as sequences, which together form the "language of nature", we introduce Nature Language Model (briefly, NatureLM), a sequence-based science foundation model designed for scientific discovery. Pre-trained with data from multiple scientific domains, NatureLM offers a unified, versatile model that enables various applications including: (i) generating and optimizing small molecules, proteins, RNA, and materials using text instructions; (ii) cross-domain generation/design, such as protein-to-molecule and protein-to-RNA generation; and (iii) achieving state-of-the-art performance in tasks like SMILES-to-IUPAC translation and retrosynthesis on USPTO-50k. NatureLM offers a promising generalist approach for various scientific tasks, including drug discovery (hit generation/optimization, ADMET optimization, synthesis), novel material design, and the development of therapeutic proteins or nucleotides. We have developed NatureLM models in different sizes (1 billion, 8 billion, and 46.7 billion parameters) and observed a clear improvement in performance as the model size increases.
Chimera: Accurate retrosynthesis prediction by ensembling models with diverse inductive biases
Dec 06, 2024



Abstract:Planning and conducting chemical syntheses remains a major bottleneck in the discovery of functional small molecules, and prevents fully leveraging generative AI for molecular inverse design. While early work has shown that ML-based retrosynthesis models can predict reasonable routes, their low accuracy for less frequent, yet important reactions has been pointed out. As multi-step search algorithms are limited to reactions suggested by the underlying model, the applicability of those tools is inherently constrained by the accuracy of retrosynthesis prediction. Inspired by how chemists use different strategies to ideate reactions, we propose Chimera: a framework for building highly accurate reaction models that combine predictions from diverse sources with complementary inductive biases using a learning-based ensembling strategy. We instantiate the framework with two newly developed models, which already by themselves achieve state of the art in their categories. Through experiments across several orders of magnitude in data scale and time-splits, we show Chimera outperforms all major models by a large margin, owing both to the good individual performance of its constituents, but also to the scalability of our ensembling strategy. Moreover, we find that PhD-level organic chemists prefer predictions from Chimera over baselines in terms of quality. Finally, we transfer the largest-scale checkpoint to an internal dataset from a major pharmaceutical company, showing robust generalization under distribution shift. With the new dimension that our framework unlocks, we anticipate further acceleration in the development of even more accurate models.
RetroGFN: Diverse and Feasible Retrosynthesis using GFlowNets
Jun 26, 2024Abstract:Single-step retrosynthesis aims to predict a set of reactions that lead to the creation of a target molecule, which is a crucial task in molecular discovery. Although a target molecule can often be synthesized with multiple different reactions, it is not clear how to verify the feasibility of a reaction, because the available datasets cover only a tiny fraction of the possible solutions. Consequently, the existing models are not encouraged to explore the space of possible reactions sufficiently. In this paper, we propose a novel single-step retrosynthesis model, RetroGFN, that can explore outside the limited dataset and return a diverse set of feasible reactions by leveraging a feasibility proxy model during the training. We show that RetroGFN achieves competitive results on standard top-k accuracy while outperforming existing methods on round-trip accuracy. Moreover, we provide empirical arguments in favor of using round-trip accuracy which expands the notion of feasibility with respect to the standard top-k accuracy metric.
Re-evaluating Retrosynthesis Algorithms with Syntheseus
Oct 30, 2023Abstract:The planning of how to synthesize molecules, also known as retrosynthesis, has been a growing focus of the machine learning and chemistry communities in recent years. Despite the appearance of steady progress, we argue that imperfect benchmarks and inconsistent comparisons mask systematic shortcomings of existing techniques. To remedy this, we present a benchmarking library called syntheseus which promotes best practice by default, enabling consistent meaningful evaluation of single-step and multi-step retrosynthesis algorithms. We use syntheseus to re-evaluate a number of previous retrosynthesis algorithms, and find that the ranking of state-of-the-art models changes when evaluated carefully. We end with guidance for future works in this area.
Retro-fallback: retrosynthetic planning in an uncertain world
Oct 13, 2023



Abstract:Retrosynthesis is the task of proposing a series of chemical reactions to create a desired molecule from simpler, buyable molecules. While previous works have proposed algorithms to find optimal solutions for a range of metrics (e.g. shortest, lowest-cost), these works generally overlook the fact that we have imperfect knowledge of the space of possible reactions, meaning plans created by the algorithm may not work in a laboratory. In this paper we propose a novel formulation of retrosynthesis in terms of stochastic processes to account for this uncertainty. We then propose a novel greedy algorithm called retro-fallback which maximizes the probability that at least one synthesis plan can be executed in the lab. Using in-silico benchmarks we demonstrate that retro-fallback generally produces better sets of synthesis plans than the popular MCTS and retro* algorithms.
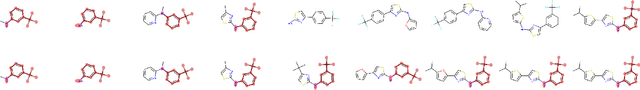
Are VAEs Bad at Reconstructing Molecular Graphs?
May 04, 2023Abstract:Many contemporary generative models of molecules are variational auto-encoders of molecular graphs. One term in their training loss pertains to reconstructing the input, yet reconstruction capabilities of state-of-the-art models have not yet been thoroughly compared on a large and chemically diverse dataset. In this work, we show that when several state-of-the-art generative models are evaluated under the same conditions, their reconstruction accuracy is surprisingly low, worse than what was previously reported on seemingly harder datasets. However, we show that improving reconstruction does not directly lead to better sampling or optimization performance. Failed reconstructions from the MoLeR model are usually similar to the inputs, assembling the same motifs in a different way, and possess similar chemical properties such as solubility. Finally, we show that the input molecule and its failed reconstruction are usually mapped by the different encoders to statistically distinguishable posterior distributions, hinting that posterior collapse may not fully explain why VAEs are bad at reconstructing molecular graphs.
Retrosynthetic Planning with Dual Value Networks
Jan 31, 2023Abstract:Retrosynthesis, which aims to find a route to synthesize a target molecule from commercially available starting materials, is a critical task in drug discovery and materials design. Recently, the combination of ML-based single-step reaction predictors with multi-step planners has led to promising results. However, the single-step predictors are mostly trained offline to optimize the single-step accuracy, without considering complete routes. Here, we leverage reinforcement learning (RL) to improve the single-step predictor, by using a tree-shaped MDP to optimize complete routes while retaining single-step accuracy. Desirable routes should be both synthesizable and of low cost. We propose an online training algorithm, called Planning with Dual Value Networks (PDVN), in which two value networks predict the synthesizability and cost of molecules, respectively. To maintain the single-step accuracy, we design a two-branch network structure for the single-step predictor. On the widely-used USPTO dataset, our PDVN algorithm improves the search success rate of existing multi-step planners (e.g., increasing the success rate from 85.79% to 98.95% for Retro*, and reducing the number of model calls by half while solving 99.47% molecules for RetroGraph). Furthermore, PDVN finds shorter synthesis routes (e.g., reducing the average route length from 5.76 to 4.83 for Retro*, and from 5.63 to 4.78 for RetroGraph).
Deep Learning for Rheumatoid Arthritis: Joint Detection and Damage Scoring in X-rays
Apr 28, 2021


Abstract:Recent advancements in computer vision promise to automate medical image analysis. Rheumatoid arthritis is an autoimmune disease that would profit from computer-based diagnosis, as there are no direct markers known, and doctors have to rely on manual inspection of X-ray images. In this work, we present a multi-task deep learning model that simultaneously learns to localize joints on X-ray images and diagnose two kinds of joint damage: narrowing and erosion. Additionally, we propose a modification of label smoothing, which combines classification and regression cues into a single loss and achieves 5% relative error reduction compared to standard loss functions. Our final model obtained 4th place in joint space narrowing and 5th place in joint erosion in the global RA2 DREAM challenge.
Learning to Extend Molecular Scaffolds with Structural Motifs
Mar 05, 2021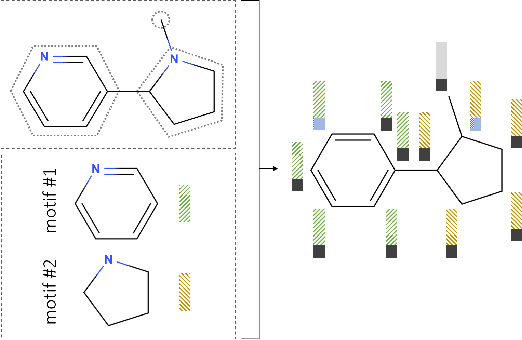
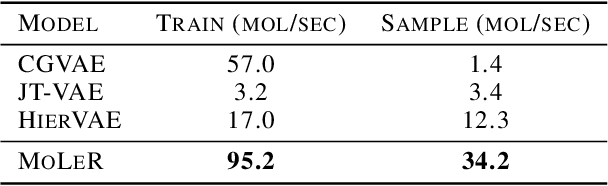
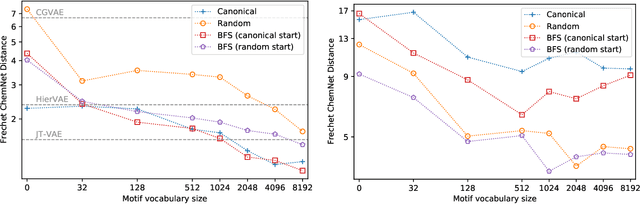
Abstract:Recent advancements in deep learning-based modeling of molecules promise to accelerate in silico drug discovery. There is a plethora of generative models available, which build molecules either atom-by-atom and bond-by-bond or fragment-by-fragment. Many drug discovery projects also require a fixed scaffold to be present in the generated molecule, and incorporating that constraint has been recently explored. In this work, we propose a new graph-based model that learns to extend a given partial molecule by flexibly choosing between adding individual atoms and entire fragments. Extending a scaffold is implemented by using it as the initial partial graph, which is possible because our model does not depend on generation history. We show that training using a randomized generation order is necessary for good performance when extending scaffolds, and that the results are further improved by increasing fragment vocabulary size. Our model pushes the state-of-the-art of graph-based molecule generation, while being an order of magnitude faster to train and sample from than existing approaches.
Holistic Multi-View Building Analysis in the Wild with Projection Pooling
Sep 25, 2020



Abstract:We address six different classification tasks related to fine-grained building attributes: construction type, number of floors, pitch and geometry of the roof, facade material, and occupancy class. Tackling such a problem of remote building analysis became possible only recently due to growing large scale datasets of urban scenes. To this end, we introduce a new benchmarking dataset, consisting of 49426 top-view and street-view images of 9674 buildings. These photos are further assembled, together with the geometric metadata. The dataset showcases a variety of real-world challenges, such as occlusions, blur, partially visible objects, and a broad spectrum of buildings. We propose a new projection pooling layer, creating a unified, top-view representation of the top-view and the side views in a high-dimensional space. It allows us to utilize the building and imagery metadata seamlessly. Introducing this layer improves classification accuracy - compared to highly tuned baseline models - indicating its suitability for building analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge