Zun Wang
OS-Symphony: A Holistic Framework for Robust and Generalist Computer-Using Agent
Jan 12, 2026Abstract:While Vision-Language Models (VLMs) have significantly advanced Computer-Using Agents (CUAs), current frameworks struggle with robustness in long-horizon workflows and generalization in novel domains. These limitations stem from a lack of granular control over historical visual context curation and the absence of visual-aware tutorial retrieval. To bridge these gaps, we introduce OS-Symphony, a holistic framework that comprises an Orchestrator coordinating two key innovations for robust automation: (1) a Reflection-Memory Agent that utilizes milestone-driven long-term memory to enable trajectory-level self-correction, effectively mitigating visual context loss in long-horizon tasks; (2) Versatile Tool Agents featuring a Multimodal Searcher that adopts a SeeAct paradigm to navigate a browser-based sandbox to synthesize live, visually aligned tutorials, thereby resolving fidelity issues in unseen scenarios. Experimental results demonstrate that OS-Symphony delivers substantial performance gains across varying model scales, establishing new state-of-the-art results on three online benchmarks, notably achieving 65.84% on OSWorld.
OS-Oracle: A Comprehensive Framework for Cross-Platform GUI Critic Models
Dec 18, 2025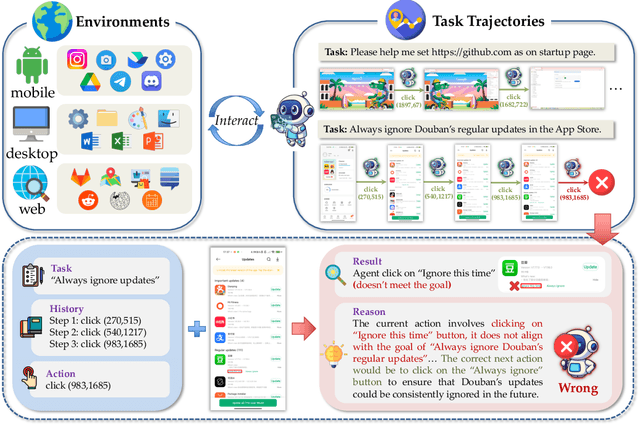
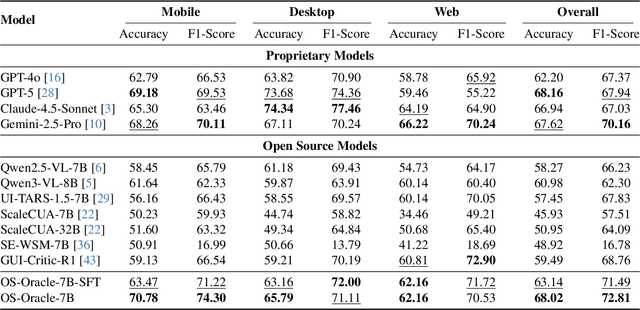
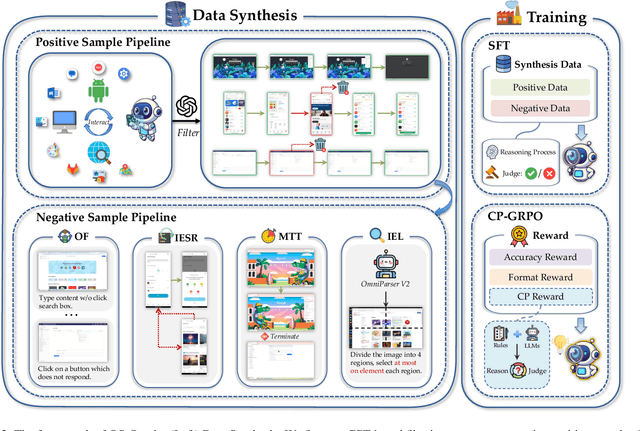
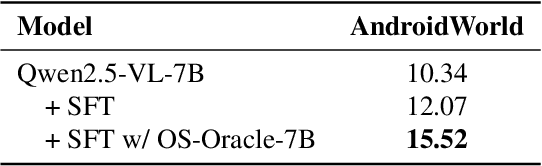
Abstract:With VLM-powered computer-using agents (CUAs) becoming increasingly capable at graphical user interface (GUI) navigation and manipulation, reliable step-level decision-making has emerged as a key bottleneck for real-world deployment. In long-horizon workflows, errors accumulate quickly and irreversible actions can cause unintended consequences, motivating critic models that assess each action before execution. While critic models offer a promising solution, their effectiveness is hindered by the lack of diverse, high-quality GUI feedback data and public critic benchmarks for step-level evaluation in computer use. To bridge these gaps, we introduce OS-Oracle that makes three core contributions: (1) a scalable data pipeline for synthesizing cross-platform GUI critic data; (2) a two-stage training paradigm combining supervised fine-tuning (SFT) and consistency-preserving group relative policy optimization (CP-GRPO); (3) OS-Critic Bench, a holistic benchmark for evaluating critic model performance across Mobile, Web, and Desktop platforms. Leveraging this framework, we curate a high-quality dataset containing 310k critic samples. The resulting critic model, OS-Oracle-7B, achieves state-of-the-art performance among open-source VLMs on OS-Critic Bench, and surpasses proprietary models on the mobile domain. Furthermore, when serving as a pre-critic, OS-Oracle-7B improves the performance of native GUI agents such as UI-TARS-1.5-7B in OSWorld and AndroidWorld environments. The code is open-sourced at https://github.com/numbmelon/OS-Oracle.
Error-Driven Scene Editing for 3D Grounding in Large Language Models
Nov 18, 2025Abstract:Despite recent progress in 3D-LLMs, they remain limited in accurately grounding language to visual and spatial elements in 3D environments. This limitation stems in part from training data that focuses on language reasoning rather than spatial understanding due to scarce 3D resources, leaving inherent grounding biases unresolved. To address this, we propose 3D scene editing as a key mechanism to generate precise visual counterfactuals that mitigate these biases through fine-grained spatial manipulation, without requiring costly scene reconstruction or large-scale 3D data collection. Furthermore, to make these edits targeted and directly address the specific weaknesses of the model, we introduce DEER-3D, an error-driven framework following a structured "Decompose, Diagnostic Evaluation, Edit, and Re-train" workflow, rather than broadly or randomly augmenting data as in conventional approaches. Specifically, upon identifying a grounding failure of the 3D-LLM, our framework first diagnoses the exact predicate-level error (e.g., attribute or spatial relation). It then executes minimal, predicate-aligned 3D scene edits, such as recoloring or repositioning, to produce targeted counterfactual supervision for iterative model fine-tuning, significantly enhancing grounding accuracy. We evaluate our editing pipeline across multiple benchmarks for 3D grounding and scene understanding tasks, consistently demonstrating improvements across all evaluated datasets through iterative refinement. DEER-3D underscores the effectiveness of targeted, error-driven scene editing in bridging linguistic reasoning capabilities with spatial grounding in 3D LLMs.
EPiC: Efficient Video Camera Control Learning with Precise Anchor-Video Guidance
May 28, 2025Abstract:Recent approaches on 3D camera control in video diffusion models (VDMs) often create anchor videos to guide diffusion models as a structured prior by rendering from estimated point clouds following annotated camera trajectories. However, errors inherent in point cloud estimation often lead to inaccurate anchor videos. Moreover, the requirement for extensive camera trajectory annotations further increases resource demands. To address these limitations, we introduce EPiC, an efficient and precise camera control learning framework that automatically constructs high-quality anchor videos without expensive camera trajectory annotations. Concretely, we create highly precise anchor videos for training by masking source videos based on first-frame visibility. This approach ensures high alignment, eliminates the need for camera trajectory annotations, and thus can be readily applied to any in-the-wild video to generate image-to-video (I2V) training pairs. Furthermore, we introduce Anchor-ControlNet, a lightweight conditioning module that integrates anchor video guidance in visible regions to pretrained VDMs, with less than 1% of backbone model parameters. By combining the proposed anchor video data and ControlNet module, EPiC achieves efficient training with substantially fewer parameters, training steps, and less data, without requiring modifications to the diffusion model backbone typically needed to mitigate rendering misalignments. Although being trained on masking-based anchor videos, our method generalizes robustly to anchor videos made with point clouds during inference, enabling precise 3D-informed camera control. EPiC achieves SOTA performance on RealEstate10K and MiraData for I2V camera control task, demonstrating precise and robust camera control ability both quantitatively and qualitatively. Notably, EPiC also exhibits strong zero-shot generalization to video-to-video scenarios.
DGRO: Enhancing LLM Reasoning via Exploration-Exploitation Control and Reward Variance Management
May 19, 2025
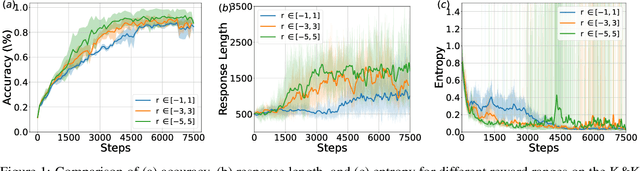
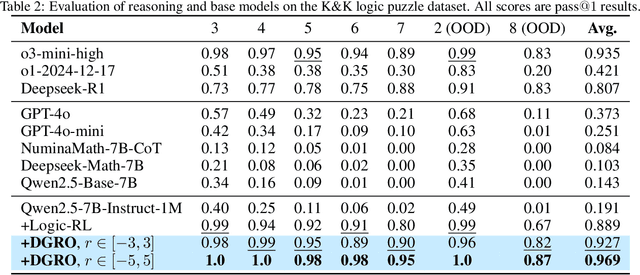
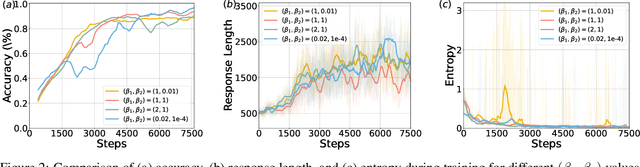
Abstract:Inference scaling further accelerates Large Language Models (LLMs) toward Artificial General Intelligence (AGI), with large-scale Reinforcement Learning (RL) to unleash long Chain-of-Thought reasoning. Most contemporary reasoning approaches usually rely on handcrafted rule-based reward functions. However, the tarde-offs of exploration and exploitation in RL algorithms involves multiple complex considerations, and the theoretical and empirical impacts of manually designed reward functions remain insufficiently explored. In this paper, we propose Decoupled Group Reward Optimization (DGRO), a general RL algorithm for LLM reasoning. On the one hand, DGRO decouples the traditional regularization coefficient into two independent hyperparameters: one scales the policy gradient term, and the other regulates the distance from the sampling policy. This decoupling not only enables precise control over balancing exploration and exploitation, but also can be seamlessly extended to Online Policy Mirror Descent (OPMD) algorithms in Kimi k1.5 and Direct Reward Optimization. On the other hand, we observe that reward variance significantly affects both convergence speed and final model performance. We conduct both theoretical analysis and extensive empirical validation to assess DGRO, including a detailed ablation study that investigates its performance and optimization dynamics. Experimental results show that DGRO achieves state-of-the-art performance on the Logic dataset with an average accuracy of 96.9\%, and demonstrates strong generalization across mathematical benchmarks.
Trust Region Preference Approximation: A simple and stable reinforcement learning algorithm for LLM reasoning
Apr 06, 2025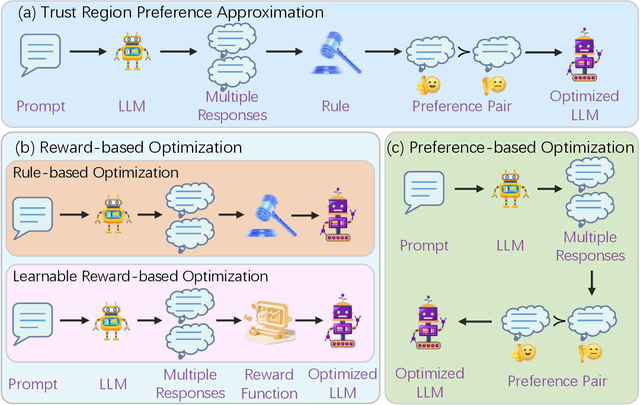
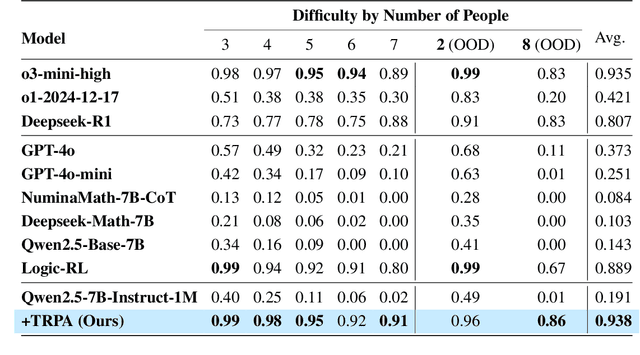
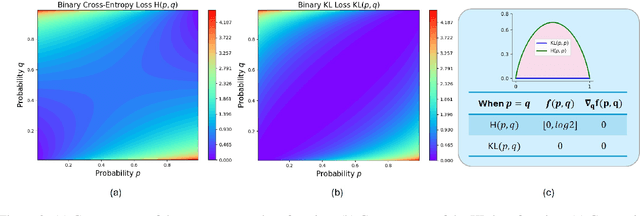
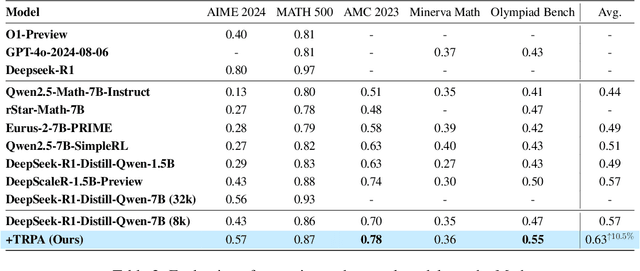
Abstract:Recently, Large Language Models (LLMs) have rapidly evolved, approaching Artificial General Intelligence (AGI) while benefiting from large-scale reinforcement learning to enhance Human Alignment (HA) and Reasoning. Recent reward-based optimization algorithms, such as Proximal Policy Optimization (PPO) and Group Relative Policy Optimization (GRPO) have achieved significant performance on reasoning tasks, whereas preference-based optimization algorithms such as Direct Preference Optimization (DPO) significantly improve the performance of LLMs on human alignment. However, despite the strong performance of reward-based optimization methods in alignment tasks , they remain vulnerable to reward hacking. Furthermore, preference-based algorithms (such as Online DPO) haven't yet matched the performance of reward-based optimization algorithms (like PPO) on reasoning tasks, making their exploration in this specific area still a worthwhile pursuit. Motivated by these challenges, we propose the Trust Region Preference Approximation (TRPA) algorithm, which integrates rule-based optimization with preference-based optimization for reasoning tasks. As a preference-based algorithm, TRPA naturally eliminates the reward hacking issue. TRPA constructs preference levels using predefined rules, forms corresponding preference pairs, and leverages a novel optimization algorithm for RL training with a theoretical monotonic improvement guarantee. Experimental results demonstrate that TRPA not only achieves competitive performance on reasoning tasks but also exhibits robust stability. The code of this paper are released and updating on https://github.com/XueruiSu/Trust-Region-Preference-Approximation.git.
Potential Score Matching: Debiasing Molecular Structure Sampling with Potential Energy Guidance
Mar 18, 2025Abstract:The ensemble average of physical properties of molecules is closely related to the distribution of molecular conformations, and sampling such distributions is a fundamental challenge in physics and chemistry. Traditional methods like molecular dynamics (MD) simulations and Markov chain Monte Carlo (MCMC) sampling are commonly used but can be time-consuming and costly. Recently, diffusion models have emerged as efficient alternatives by learning the distribution of training data. Obtaining an unbiased target distribution is still an expensive task, primarily because it requires satisfying ergodicity. To tackle these challenges, we propose Potential Score Matching (PSM), an approach that utilizes the potential energy gradient to guide generative models. PSM does not require exact energy functions and can debias sample distributions even when trained on limited and biased data. Our method outperforms existing state-of-the-art (SOTA) models on the Lennard-Jones (LJ) potential, a commonly used toy model. Furthermore, we extend the evaluation of PSM to high-dimensional problems using the MD17 and MD22 datasets. The results demonstrate that molecular distributions generated by PSM more closely approximate the Boltzmann distribution compared to traditional diffusion models.
UniGenX: Unified Generation of Sequence and Structure with Autoregressive Diffusion
Mar 09, 2025Abstract:Unified generation of sequence and structure for scientific data (e.g., materials, molecules, proteins) is a critical task. Existing approaches primarily rely on either autoregressive sequence models or diffusion models, each offering distinct advantages and facing notable limitations. Autoregressive models, such as GPT, Llama, and Phi-4, have demonstrated remarkable success in natural language generation and have been extended to multimodal tasks (e.g., image, video, and audio) using advanced encoders like VQ-VAE to represent complex modalities as discrete sequences. However, their direct application to scientific domains is challenging due to the high precision requirements and the diverse nature of scientific data. On the other hand, diffusion models excel at generating high-dimensional scientific data, such as protein, molecule, and material structures, with remarkable accuracy. Yet, their inability to effectively model sequences limits their potential as general-purpose multimodal foundation models. To address these challenges, we propose UniGenX, a unified framework that combines autoregressive next-token prediction with conditional diffusion models. This integration leverages the strengths of autoregressive models to ease the training of conditional diffusion models, while diffusion-based generative heads enhance the precision of autoregressive predictions. We validate the effectiveness of UniGenX on material and small molecule generation tasks, achieving a significant leap in state-of-the-art performance for material crystal structure prediction and establishing new state-of-the-art results for small molecule structure prediction, de novo design, and conditional generation. Notably, UniGenX demonstrates significant improvements, especially in handling long sequences for complex structures, showcasing its efficacy as a versatile tool for scientific data generation.
Enhancing the Scalability and Applicability of Kohn-Sham Hamiltonians for Molecular Systems
Feb 26, 2025Abstract:Density Functional Theory (DFT) is a pivotal method within quantum chemistry and materials science, with its core involving the construction and solution of the Kohn-Sham Hamiltonian. Despite its importance, the application of DFT is frequently limited by the substantial computational resources required to construct the Kohn-Sham Hamiltonian. In response to these limitations, current research has employed deep-learning models to efficiently predict molecular and solid Hamiltonians, with roto-translational symmetries encoded in their neural networks. However, the scalability of prior models may be problematic when applied to large molecules, resulting in non-physical predictions of ground-state properties. In this study, we generate a substantially larger training set (PubChemQH) than used previously and use it to create a scalable model for DFT calculations with physical accuracy. For our model, we introduce a loss function derived from physical principles, which we call Wavefunction Alignment Loss (WALoss). WALoss involves performing a basis change on the predicted Hamiltonian to align it with the observed one; thus, the resulting differences can serve as a surrogate for orbital energy differences, allowing models to make better predictions for molecular orbitals and total energies than previously possible. WALoss also substantially accelerates self-consistent-field (SCF) DFT calculations. Here, we show it achieves a reduction in total energy prediction error by a factor of 1347 and an SCF calculation speed-up by a factor of 18%. These substantial improvements set new benchmarks for achieving accurate and applicable predictions in larger molecular systems.
NatureLM: Deciphering the Language of Nature for Scientific Discovery
Feb 11, 2025



Abstract:Foundation models have revolutionized natural language processing and artificial intelligence, significantly enhancing how machines comprehend and generate human languages. Inspired by the success of these foundation models, researchers have developed foundation models for individual scientific domains, including small molecules, materials, proteins, DNA, and RNA. However, these models are typically trained in isolation, lacking the ability to integrate across different scientific domains. Recognizing that entities within these domains can all be represented as sequences, which together form the "language of nature", we introduce Nature Language Model (briefly, NatureLM), a sequence-based science foundation model designed for scientific discovery. Pre-trained with data from multiple scientific domains, NatureLM offers a unified, versatile model that enables various applications including: (i) generating and optimizing small molecules, proteins, RNA, and materials using text instructions; (ii) cross-domain generation/design, such as protein-to-molecule and protein-to-RNA generation; and (iii) achieving state-of-the-art performance in tasks like SMILES-to-IUPAC translation and retrosynthesis on USPTO-50k. NatureLM offers a promising generalist approach for various scientific tasks, including drug discovery (hit generation/optimization, ADMET optimization, synthesis), novel material design, and the development of therapeutic proteins or nucleotides. We have developed NatureLM models in different sizes (1 billion, 8 billion, and 46.7 billion parameters) and observed a clear improvement in performance as the model size increases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge