Lin Huang
Elign: Equivariant Diffusion Model Alignment from Foundational Machine Learning Force Fields
Jan 29, 2026Abstract:Generative models for 3D molecular conformations must respect Euclidean symmetries and concentrate probability mass on thermodynamically favorable, mechanically stable structures. However, E(3)-equivariant diffusion models often reproduce biases from semi-empirical training data rather than capturing the equilibrium distribution of a high-fidelity Hamiltonian. While physics-based guidance can correct this, it faces two computational bottlenecks: expensive quantum-chemical evaluations (e.g., DFT) and the need to repeat such queries at every sampling step. We present Elign, a post-training framework that amortizes both costs. First, we replace expensive DFT evaluations with a faster, pretrained foundational machine-learning force field (MLFF) to provide physical signals. Second, we eliminate repeated run-time queries by shifting physical steering to the training phase. To achieve the second amortization, we formulate reverse diffusion as a reinforcement learning problem and introduce Force--Energy Disentangled Group Relative Policy Optimization (FED-GRPO) to fine-tune the denoising policy. FED-GRPO includes a potential-based energy reward and a force-based stability reward, which are optimized and group-normalized independently. Experiments show that Elign generates conformations with lower gold-standard DFT energies and forces, while improving stability. Crucially, inference remains as fast as unguided sampling, since no energy evaluations are required during generation.
YOLO-DS: Fine-Grained Feature Decoupling via Dual-Statistic Synergy Operator for Object Detection
Jan 26, 2026Abstract:One-stage object detection, particularly the YOLO series, strikes a favorable balance between accuracy and efficiency. However, existing YOLO detectors lack explicit modeling of heterogeneous object responses within shared feature channels, which limits further performance gains. To address this, we propose YOLO-DS, a framework built around a novel Dual-Statistic Synergy Operator (DSO). The DSO decouples object features by jointly modeling the channel-wise mean and the peak-to-mean difference. Building upon the DSO, we design two lightweight gating modules: the Dual-Statistic Synergy Gating (DSG) module for adaptive channel-wise feature selection, and the Multi-Path Segmented Gating (MSG) module for depth-wise feature weighting. On the MS-COCO benchmark, YOLO-DS consistently outperforms YOLOv8 across five model scales (N, S, M, L, X), achieving AP gains of 1.1% to 1.7% with only a minimal increase in inference latency. Extensive visualization, ablation, and comparative studies validate the effectiveness of our approach, demonstrating its superior capability in discriminating heterogeneous objects with high efficiency.
E2Former-V2: On-the-Fly Equivariant Attention with Linear Activation Memory
Jan 23, 2026Abstract:Equivariant Graph Neural Networks (EGNNs) have become a widely used approach for modeling 3D atomistic systems. However, mainstream architectures face critical scalability bottlenecks due to the explicit construction of geometric features or dense tensor products on \textit{every} edge. To overcome this, we introduce \textbf{E2Former-V2}, a scalable architecture that integrates algebraic sparsity with hardware-aware execution. We first propose \textbf{E}quivariant \textbf{A}xis-\textbf{A}ligned \textbf{S}parsification (EAAS). EAAS builds on Wigner-$6j$ convolution by exploiting an $\mathrm{SO}(3) \rightarrow \mathrm{SO}(2)$ change of basis to transform computationally expensive dense tensor contractions into efficient, sparse parity re-indexing operations. Building on this representation, we introduce \textbf{On-the-Fly Equivariant Attention}, a fully node-centric mechanism implemented via a custom fused Triton kernel. By eliminating materialized edge tensors and maximizing SRAM utilization, our kernel achieves a \textbf{20$\times$ improvement in TFLOPS} compared to standard implementations. Extensive experiments on the SPICE and OMol25 datasets demonstrate that E2Former-V2 maintains comparable predictive performance while notably accelerating inference. This work demonstrates that large equivariant transformers can be trained efficiently using widely accessible GPU platforms. The code is avalible at https://github.com/IQuestLab/UBio-MolFM/tree/e2formerv2.
Scalable Machine Learning Force Fields for Macromolecular Systems Through Long-Range Aware Message Passing
Jan 07, 2026Abstract:Machine learning force fields (MLFFs) have revolutionized molecular simulations by providing quantum mechanical accuracy at the speed of molecular mechanical computations. However, a fundamental reliance of these models on fixed-cutoff architectures limits their applicability to macromolecular systems where long-range interactions dominate. We demonstrate that this locality constraint causes force prediction errors to scale monotonically with system size, revealing a critical architectural bottleneck. To overcome this, we establish the systematically designed MolLR25 ({Mol}ecules with {L}ong-{R}ange effect) benchmark up to 1200 atoms, generated using high-fidelity DFT, and introduce E2Former-LSR, an equivariant transformer that explicitly integrates long-range attention blocks. E2Former-LSR exhibits stable error scaling, achieves superior fidelity in capturing non-covalent decay, and maintains precision on complex protein conformations. Crucially, its efficient design provides up to 30% speedup compared to purely local models. This work validates the necessity of non-local architectures for generalizable MLFFs, enabling high-fidelity molecular dynamics for large-scale chemical and biological systems.
Accurate and scalable exchange-correlation with deep learning
Jun 18, 2025Abstract:Density Functional Theory (DFT) is the most widely used electronic structure method for predicting the properties of molecules and materials. Although DFT is, in principle, an exact reformulation of the Schr\"odinger equation, practical applications rely on approximations to the unknown exchange-correlation (XC) functional. Most existing XC functionals are constructed using a limited set of increasingly complex, hand-crafted features that improve accuracy at the expense of computational efficiency. Yet, no current approximation achieves the accuracy and generality for predictive modeling of laboratory experiments at chemical accuracy -- typically defined as errors below 1 kcal/mol. In this work, we present Skala, a modern deep learning-based XC functional that bypasses expensive hand-designed features by learning representations directly from data. Skala achieves chemical accuracy for atomization energies of small molecules while retaining the computational efficiency typical of semi-local DFT. This performance is enabled by training on an unprecedented volume of high-accuracy reference data generated using computationally intensive wavefunction-based methods. Notably, Skala systematically improves with additional training data covering diverse chemistry. By incorporating a modest amount of additional high-accuracy data tailored to chemistry beyond atomization energies, Skala achieves accuracy competitive with the best-performing hybrid functionals across general main group chemistry, at the cost of semi-local DFT. As the training dataset continues to expand, Skala is poised to further enhance the predictive power of first-principles simulations.
YOLO-PRO: Enhancing Instance-Specific Object Detection with Full-Channel Global Self-Attention
Mar 04, 2025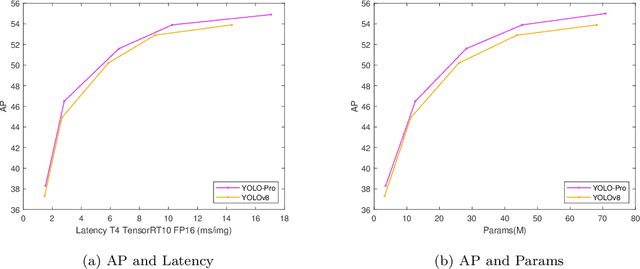
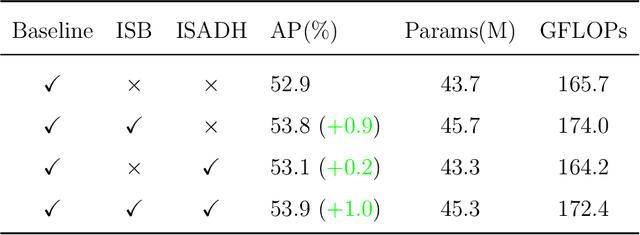
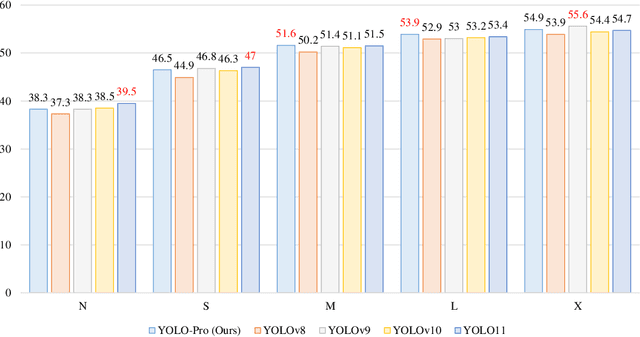
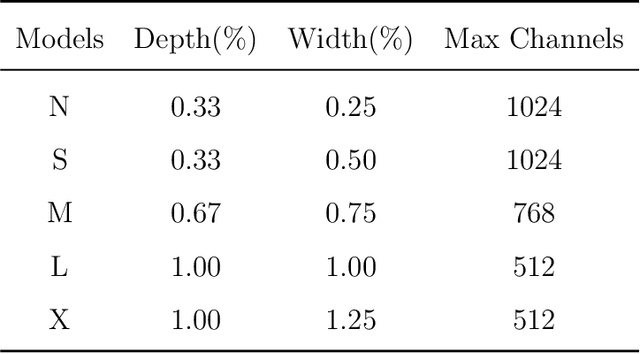
Abstract:This paper addresses the inherent limitations of conventional bottleneck structures (diminished instance discriminability due to overemphasis on batch statistics) and decoupled heads (computational redundancy) in object detection frameworks by proposing two novel modules: the Instance-Specific Bottleneck with full-channel global self-attention (ISB) and the Instance-Specific Asymmetric Decoupled Head (ISADH). The ISB module innovatively reconstructs feature maps to establish an efficient full-channel global attention mechanism through synergistic fusion of batch-statistical and instance-specific features. Complementing this, the ISADH module pioneers an asymmetric decoupled architecture enabling hierarchical multi-dimensional feature integration via dual-stream batch-instance representation fusion. Extensive experiments on the MS-COCO benchmark demonstrate that the coordinated deployment of ISB and ISADH in the YOLO-PRO framework achieves state-of-the-art performance across all computational scales. Specifically, YOLO-PRO surpasses YOLOv8 by 1.0-1.6% AP (N/S/M/L/X scales) and outperforms YOLO11 by 0.1-0.5% AP in critical M/L/X groups, while maintaining competitive computational efficiency. This work provides practical insights for developing high-precision detectors deployable on edge devices.
Enhancing the Scalability and Applicability of Kohn-Sham Hamiltonians for Molecular Systems
Feb 26, 2025Abstract:Density Functional Theory (DFT) is a pivotal method within quantum chemistry and materials science, with its core involving the construction and solution of the Kohn-Sham Hamiltonian. Despite its importance, the application of DFT is frequently limited by the substantial computational resources required to construct the Kohn-Sham Hamiltonian. In response to these limitations, current research has employed deep-learning models to efficiently predict molecular and solid Hamiltonians, with roto-translational symmetries encoded in their neural networks. However, the scalability of prior models may be problematic when applied to large molecules, resulting in non-physical predictions of ground-state properties. In this study, we generate a substantially larger training set (PubChemQH) than used previously and use it to create a scalable model for DFT calculations with physical accuracy. For our model, we introduce a loss function derived from physical principles, which we call Wavefunction Alignment Loss (WALoss). WALoss involves performing a basis change on the predicted Hamiltonian to align it with the observed one; thus, the resulting differences can serve as a surrogate for orbital energy differences, allowing models to make better predictions for molecular orbitals and total energies than previously possible. WALoss also substantially accelerates self-consistent-field (SCF) DFT calculations. Here, we show it achieves a reduction in total energy prediction error by a factor of 1347 and an SCF calculation speed-up by a factor of 18%. These substantial improvements set new benchmarks for achieving accurate and applicable predictions in larger molecular systems.
Efficient and Scalable Density Functional Theory Hamiltonian Prediction through Adaptive Sparsity
Feb 03, 2025Abstract:Hamiltonian matrix prediction is pivotal in computational chemistry, serving as the foundation for determining a wide range of molecular properties. While SE(3) equivariant graph neural networks have achieved remarkable success in this domain, their substantial computational cost-driven by high-order tensor product (TP) operations-restricts their scalability to large molecular systems with extensive basis sets. To address this challenge, we introduce SPHNet, an efficient and scalable equivariant network that incorporates adaptive sparsity into Hamiltonian prediction. SPHNet employs two innovative sparse gates to selectively constrain non-critical interaction combinations, significantly reducing tensor product computations while maintaining accuracy. To optimize the sparse representation, we develop a Three-phase Sparsity Scheduler, ensuring stable convergence and achieving high performance at sparsity rates of up to 70 percent. Extensive evaluations on QH9 and PubchemQH datasets demonstrate that SPHNet achieves state-of-the-art accuracy while providing up to a 7x speedup over existing models. Beyond Hamiltonian prediction, the proposed sparsification techniques also hold significant potential for improving the efficiency and scalability of other SE(3) equivariant networks, further broadening their applicability and impact.
\underline{E2}Former: A Linear-time \underline{E}fficient and \underline{E}quivariant Trans\underline{former} for Scalable Molecular Modeling
Jan 31, 2025



Abstract:Equivariant Graph Neural Networks (EGNNs) have demonstrated significant success in modeling microscale systems, including those in chemistry, biology and materials science. However, EGNNs face substantial computational challenges due to the high cost of constructing edge features via spherical tensor products, making them impractical for large-scale systems. To address this limitation, we introduce E2Former, an equivariant and efficient transformer architecture that incorporates the Wigner $6j$ convolution (Wigner $6j$ Conv). By shifting the computational burden from edges to nodes, the Wigner $6j$ Conv reduces the complexity from $O(|\mathcal{E}|)$ to $ O(| \mathcal{V}|)$ while preserving both the model's expressive power and rotational equivariance. We show that this approach achieves a 7x-30x speedup compared to conventional $\mathrm{SO}(3)$ convolutions. Furthermore, our empirical results demonstrate that the derived E2Former mitigates the computational challenges of existing approaches without compromising the ability to capture detailed geometric information. This development could suggest a promising direction for scalable and efficient molecular modeling.
Physical Consistency Bridges Heterogeneous Data in Molecular Multi-Task Learning
Oct 14, 2024



Abstract:In recent years, machine learning has demonstrated impressive capability in handling molecular science tasks. To support various molecular properties at scale, machine learning models are trained in the multi-task learning paradigm. Nevertheless, data of different molecular properties are often not aligned: some quantities, e.g. equilibrium structure, demand more cost to compute than others, e.g. energy, so their data are often generated by cheaper computational methods at the cost of lower accuracy, which cannot be directly overcome through multi-task learning. Moreover, it is not straightforward to leverage abundant data of other tasks to benefit a particular task. To handle such data heterogeneity challenges, we exploit the specialty of molecular tasks that there are physical laws connecting them, and design consistency training approaches that allow different tasks to exchange information directly so as to improve one another. Particularly, we demonstrate that the more accurate energy data can improve the accuracy of structure prediction. We also find that consistency training can directly leverage force and off-equilibrium structure data to improve structure prediction, demonstrating a broad capability for integrating heterogeneous data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge