Dihan Zheng
Enhancing Low-resolution Image Representation Through Normalizing Flows
Jan 11, 2026Abstract:Low-resolution image representation is a special form of sparse representation that retains only low-frequency information while discarding high-frequency components. This property reduces storage and transmission costs and benefits various image processing tasks. However, a key challenge is to preserve essential visual content while maintaining the ability to accurately reconstruct the original images. This work proposes LR2Flow, a nonlinear framework that learns low-resolution image representations by integrating wavelet tight frame blocks with normalizing flows. We conduct a reconstruction error analysis of the proposed network, which demonstrates the necessity of designing invertible neural networks in the wavelet tight frame domain. Experimental results on various tasks, including image rescaling, compression, and denoising, demonstrate the effectiveness of the learned representations and the robustness of the proposed framework.
Physical Consistency Bridges Heterogeneous Data in Molecular Multi-Task Learning
Oct 14, 2024



Abstract:In recent years, machine learning has demonstrated impressive capability in handling molecular science tasks. To support various molecular properties at scale, machine learning models are trained in the multi-task learning paradigm. Nevertheless, data of different molecular properties are often not aligned: some quantities, e.g. equilibrium structure, demand more cost to compute than others, e.g. energy, so their data are often generated by cheaper computational methods at the cost of lower accuracy, which cannot be directly overcome through multi-task learning. Moreover, it is not straightforward to leverage abundant data of other tasks to benefit a particular task. To handle such data heterogeneity challenges, we exploit the specialty of molecular tasks that there are physical laws connecting them, and design consistency training approaches that allow different tasks to exchange information directly so as to improve one another. Particularly, we demonstrate that the more accurate energy data can improve the accuracy of structure prediction. We also find that consistency training can directly leverage force and off-equilibrium structure data to improve structure prediction, demonstrating a broad capability for integrating heterogeneous data.
SeNM-VAE: Semi-Supervised Noise Modeling with Hierarchical Variational Autoencoder
Mar 26, 2024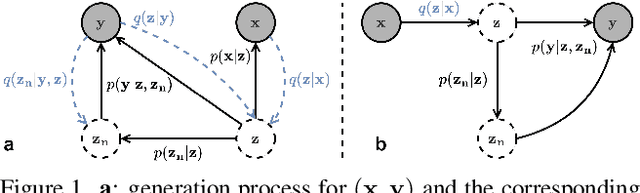
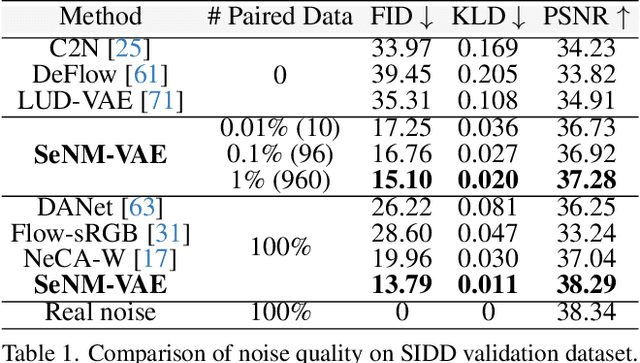
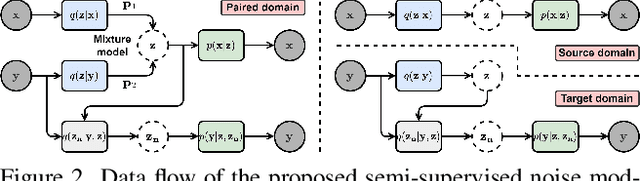
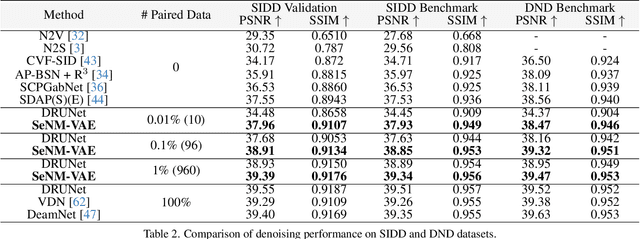
Abstract:The data bottleneck has emerged as a fundamental challenge in learning based image restoration methods. Researchers have attempted to generate synthesized training data using paired or unpaired samples to address this challenge. This study proposes SeNM-VAE, a semi-supervised noise modeling method that leverages both paired and unpaired datasets to generate realistic degraded data. Our approach is based on modeling the conditional distribution of degraded and clean images with a specially designed graphical model. Under the variational inference framework, we develop an objective function for handling both paired and unpaired data. We employ our method to generate paired training samples for real-world image denoising and super-resolution tasks. Our approach excels in the quality of synthetic degraded images compared to other unpaired and paired noise modeling methods. Furthermore, our approach demonstrates remarkable performance in downstream image restoration tasks, even with limited paired data. With more paired data, our method achieves the best performance on the SIDD dataset.
Addressing preferred orientation in single-particle cryo-EM through AI-generated auxiliary particles
Sep 26, 2023Abstract:The single-particle cryo-EM field faces the persistent challenge of preferred orientation, lacking general computational solutions. We introduce cryoPROS, an AI-based approach designed to address the above issue. By generating the auxiliary particles with a conditional deep generative model, cryoPROS addresses the intrinsic bias in orientation estimation for the observed particles. We effectively employed cryoPROS in the cryo-EM single particle analysis of the hemagglutinin trimer, showing the ability to restore the near-atomic resolution structure on non-tilt data. Moreover, the enhanced version named cryoPROS-MP significantly improves the resolution of the membrane protein NaX using the no-tilted data that contains the effects of micelles. Compared to the classical approaches, cryoPROS does not need special experimental or image acquisition techniques, providing a purely computational yet effective solution for the preferred orientation problem. Finally, we conduct extensive experiments that establish the low risk of model bias and the high robustness of cryoPROS.
Learn from Unpaired Data for Image Restoration: A Variational Bayes Approach
Apr 21, 2022



Abstract:Collecting paired training data is difficult in practice, but the unpaired samples broadly exist. Current approaches aim at generating synthesized training data from the unpaired samples by exploring the relationship between the corrupted and clean data. This work proposes LUD-VAE, a deep generative method to learn the joint probability density function from data sampled from marginal distributions. Our approach is based on a carefully designed probabilistic graphical model in which the clean and corrupted data domains are conditionally independent. Using variational inference, we maximize the evidence lower bound (ELBO) to estimate the joint probability density function. Furthermore, we show that the ELBO is computable without paired samples under the inference invariant assumption. This property provides the mathematical rationale of our approach in the unpaired setting. Finally, we apply our method to real-world image denoising and super-resolution tasks and train the models using the synthetic data generated by the LUD-VAE. Experimental results validate the advantages of our method over other learnable approaches.
Unsupervised Deep Learning Meets Chan-Vese Model
Apr 14, 2022



Abstract:The Chan-Vese (CV) model is a classic region-based method in image segmentation. However, its piecewise constant assumption does not always hold for practical applications. Many improvements have been proposed but the issue is still far from well solved. In this work, we propose an unsupervised image segmentation approach that integrates the CV model with deep neural networks, which significantly improves the original CV model's segmentation accuracy. Our basic idea is to apply a deep neural network that maps the image into a latent space to alleviate the violation of the piecewise constant assumption in image space. We formulate this idea under the classic Bayesian framework by approximating the likelihood with an evidence lower bound (ELBO) term while keeping the prior term in the CV model. Thus, our model only needs the input image itself and does not require pre-training from external datasets. Moreover, we extend the idea to multi-phase case and dataset based unsupervised image segmentation. Extensive experiments validate the effectiveness of our model and show that the proposed method is noticeably better than other unsupervised segmentation approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge