Xinzhu Ma
GraspLDP: Towards Generalizable Grasping Policy via Latent Diffusion
Feb 26, 2026Abstract:This paper focuses on enhancing the grasping precision and generalization of manipulation policies learned via imitation learning. Diffusion-based policy learning methods have recently become the mainstream approach for robotic manipulation tasks. As grasping is a critical subtask in manipulation, the ability of imitation-learned policies to execute precise and generalizable grasps merits particular attention. Existing imitation learning techniques for grasping often suffer from imprecise grasp executions, limited spatial generalization, and poor object generalization. To address these challenges, we incorporate grasp prior knowledge into the diffusion policy framework. In particular, we employ a latent diffusion policy to guide action chunk decoding with grasp pose prior, ensuring that generated motion trajectories adhere closely to feasible grasp configurations. Furthermore, we introduce a self-supervised reconstruction objective during diffusion to embed the graspness prior: at each reverse diffusion step, we reconstruct wrist-camera images back-projected the graspness from the intermediate representations. Both simulation and real robot experiments demonstrate that our approach significantly outperforms baseline methods and exhibits strong dynamic grasping capabilities.
Charting Empirical Laws for LLM Fine-Tuning in Scientific Multi-Discipline Learning
Feb 11, 2026Abstract:While large language models (LLMs) have achieved strong performance through fine-tuning within individual scientific domains, their learning dynamics in multi-disciplinary contexts remains poorly understood, despite the promise of improved generalization and broader applicability through cross-domain knowledge synergy. In this work, we present the first systematic study of multi-disciplinary LLM fine-tuning, constructing a five-discipline corpus and analyzing learning patterns of full fine-tuning, LoRA, LoRA-MoE, and LoRA compositions. Particularly, our study shows that multi-disciplinary learning is substantially more variable than single-discipline training and distills four consistent empirical laws: (1) Balance-then-Diversity: low-resource disciplines degrade performance unless mitigated via diversity-aware upsampling; (2) Merge-then-Align: restoring instruction-following ability is critical for cross-discipline synergy; (3) Optimize-then-Scale: parameter scaling offers limited gains without prior design optimization; and (4) Share-then-Specialize: asymmetric LoRA-MoE yields robust gains with minimal trainable parameters via shared low-rank projection. Together, these laws form a practical recipe for principled multi-discipline fine-tuning and provide actionable guidance for developing generalizable scientific LLMs.
Towards Unbiased Source-Free Object Detection via Vision Foundation Models
Jan 19, 2026Abstract:Source-Free Object Detection (SFOD) has garnered much attention in recent years by eliminating the need of source-domain data in cross-domain tasks, but existing SFOD methods suffer from the Source Bias problem, i.e. the adapted model remains skewed towards the source domain, leading to poor generalization and error accumulation during self-training. To overcome this challenge, we propose Debiased Source-free Object Detection (DSOD), a novel VFM-assisted SFOD framework that can effectively mitigate source bias with the help of powerful VFMs. Specifically, we propose Unified Feature Injection (UFI) module that integrates VFM features into the CNN backbone through Simple-Scale Extension (SSE) and Domain-aware Adaptive Weighting (DAAW). Then, we propose Semantic-aware Feature Regularization (SAFR) that constrains feature learning to prevent overfitting to source domain characteristics. Furthermore, we propose a VFM-free variant, termed DSOD-distill for computation-restricted scenarios through a novel Dual-Teacher distillation scheme. Extensive experiments on multiple benchmarks demonstrate that DSOD outperforms state-of-the-art SFOD methods, achieving 48.1% AP on Normal-to-Foggy weather adaptation, 39.3% AP on Cross-scene adaptation, and 61.4% AP on Synthetic-to-Real adaptation.
SciIF: Benchmarking Scientific Instruction Following Towards Rigorous Scientific Intelligence
Jan 08, 2026Abstract:As large language models (LLMs) transition from general knowledge retrieval to complex scientific discovery, their evaluation standards must also incorporate the rigorous norms of scientific inquiry. Existing benchmarks exhibit a critical blind spot: general instruction-following metrics focus on superficial formatting, while domain-specific scientific benchmarks assess only final-answer correctness, often rewarding models that arrive at the right result with the wrong reasons. To address this gap, we introduce scientific instruction following: the capability to solve problems while strictly adhering to the constraints that establish scientific validity. Specifically, we introduce SciIF, a multi-discipline benchmark that evaluates this capability by pairing university-level problems with a fixed catalog of constraints across three pillars: scientific conditions (e.g., boundary checks and assumptions), semantic stability (e.g., unit and symbol conventions), and specific processes(e.g., required numerical methods). Uniquely, SciIF emphasizes auditability, requiring models to provide explicit evidence of constraint satisfaction rather than implicit compliance. By measuring both solution correctness and multi-constraint adherence, SciIF enables finegrained diagnosis of compositional reasoning failures, ensuring that LLMs can function as reliable agents within the strict logical frameworks of science.
Propagating Sparse Depth via Depth Foundation Model for Out-of-Distribution Depth Completion
Aug 07, 2025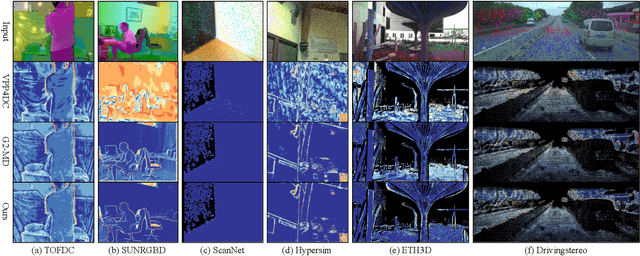
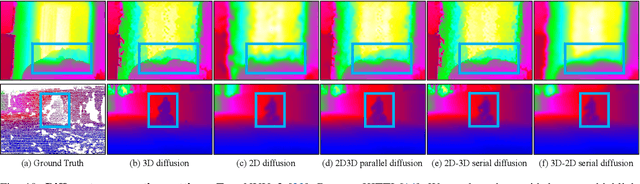
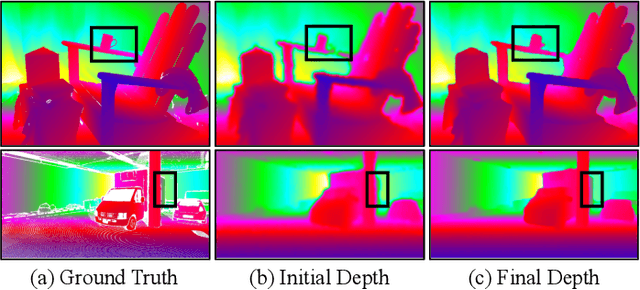
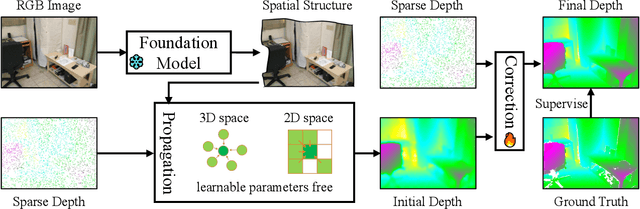
Abstract:Depth completion is a pivotal challenge in computer vision, aiming at reconstructing the dense depth map from a sparse one, typically with a paired RGB image. Existing learning based models rely on carefully prepared but limited data, leading to significant performance degradation in out-of-distribution (OOD) scenarios. Recent foundation models have demonstrated exceptional robustness in monocular depth estimation through large-scale training, and using such models to enhance the robustness of depth completion models is a promising solution. In this work, we propose a novel depth completion framework that leverages depth foundation models to attain remarkable robustness without large-scale training. Specifically, we leverage a depth foundation model to extract environmental cues, including structural and semantic context, from RGB images to guide the propagation of sparse depth information into missing regions. We further design a dual-space propagation approach, without any learnable parameters, to effectively propagates sparse depth in both 3D and 2D spaces to maintain geometric structure and local consistency. To refine the intricate structure, we introduce a learnable correction module to progressively adjust the depth prediction towards the real depth. We train our model on the NYUv2 and KITTI datasets as in-distribution datasets and extensively evaluate the framework on 16 other datasets. Our framework performs remarkably well in the OOD scenarios and outperforms existing state-of-the-art depth completion methods. Our models are released in https://github.com/shenglunch/PSD.
SP-VLA: A Joint Model Scheduling and Token Pruning Approach for VLA Model Acceleration
Jun 15, 2025Abstract:Vision-Language-Action (VLA) models have attracted increasing attention for their strong control capabilities. However, their high computational cost and low execution frequency hinder their suitability for real-time tasks such as robotic manipulation and autonomous navigation. Existing VLA acceleration methods primarily focus on structural optimization, overlooking the fact that these models operate in sequential decision-making environments. As a result, temporal redundancy in sequential action generation and spatial redundancy in visual input remain unaddressed. To this end, we propose SP-VLA, a unified framework that accelerates VLA models by jointly scheduling models and pruning tokens. Specifically, we design an action-aware model scheduling mechanism that reduces temporal redundancy by dynamically switching between VLA model and a lightweight generator. Inspired by the human motion pattern of focusing on key decision points while relying on intuition for other actions, we categorize VLA actions into deliberative and intuitive, assigning the former to the VLA model and the latter to the lightweight generator, enabling frequency-adaptive execution through collaborative model scheduling. To address spatial redundancy, we further develop a spatio-semantic dual-aware token pruning method. Tokens are classified into spatial and semantic types and pruned based on their dual-aware importance to accelerate VLA inference. These two mechanisms work jointly to guide the VLA in focusing on critical actions and salient visual information, achieving effective acceleration while maintaining high accuracy. Experimental results demonstrate that our method achieves up to 1.5$\times$ acceleration with less than 3% drop in accuracy, outperforming existing approaches in multiple tasks.
CPRet: A Dataset, Benchmark, and Model for Retrieval in Competitive Programming
May 19, 2025Abstract:Competitive programming benchmarks are widely used in scenarios such as programming contests and large language model assessments. However, the growing presence of duplicate or highly similar problems raises concerns not only about competition fairness, but also about the validity of competitive programming as a benchmark for model evaluation. In this paper, we propose a new problem -- similar question retrieval -- to address this issue. Due to the lack of both data and models, solving this problem is challenging. To this end, we introduce CPRet, a retrieval-oriented benchmark suite for competitive programming, covering four retrieval tasks: two code-centric (i.e., Text-to-Code and Code-to-Code) and two newly proposed problem-centric tasks (i.e., Problem-to-Duplicate and Simplified-to-Full), built from a combination of automatically crawled problem-solution data and manually curated annotations. Our contribution includes both high-quality training data and temporally separated test sets for reliable evaluation. In addition, we develop two task-specialized retrievers based on this dataset: CPRetriever-Code, trained with a novel Group-InfoNCE loss for problem-code alignment, and CPRetriever-Prob, fine-tuned for identifying problem-level similarity. Both models achieve strong results and are open-sourced for local use. Finally, we analyze LiveCodeBench and find that high-similarity problems inflate model pass rates and reduce differentiation, underscoring the need for similarity-aware evaluation in future benchmarks. Code and data are available at: https://github.com/coldchair/CPRet
Point2Primitive: CAD Reconstruction from Point Cloud by Direct Primitive Prediction
May 04, 2025
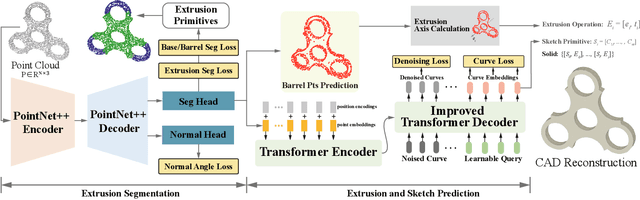
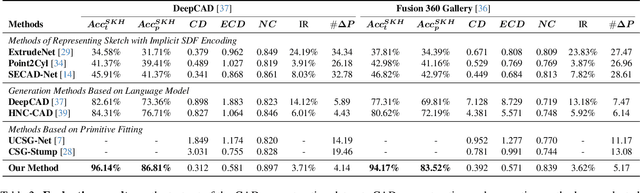
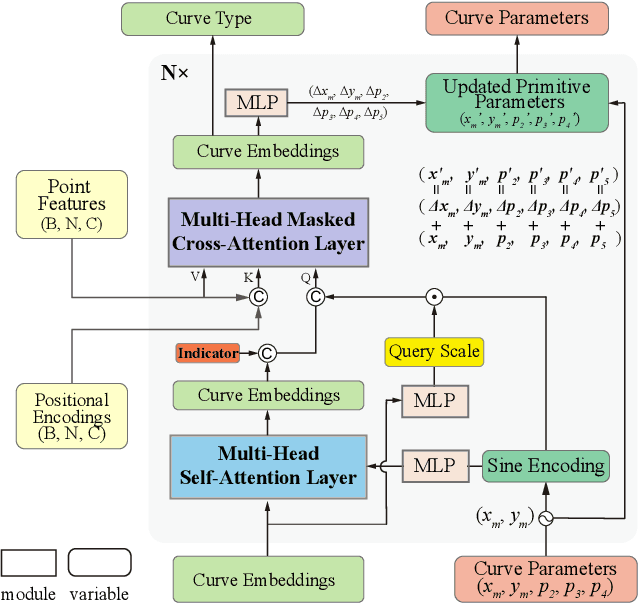
Abstract:Recovering CAD models from point clouds, especially the sketch-extrusion process, can be seen as the process of rebuilding the topology and extrusion primitives. Previous methods utilize implicit fields for sketch representation, leading to shape reconstruction of curved edges. In this paper, we proposed a CAD reconstruction network that produces editable CAD models from input point clouds (Point2Primitive) by directly predicting every element of the extrusion primitives. Point2Primitive can directly detect and predict sketch curves (type and parameter) from point clouds based on an improved transformer. The sketch curve parameters are formulated as position queries and optimized in an autoregressive way, leading to high parameter accuracy. The topology is rebuilt by extrusion segmentation, and each extrusion parameter (sketch and extrusion operation) is recovered by combining the predicted curves and the computed extrusion operation. Extensive experiments demonstrate that our method is superior in primitive prediction accuracy and CAD reconstruction. The reconstructed shapes are of high geometrical fidelity.
CMT: A Cascade MAR with Topology Predictor for Multimodal Conditional CAD Generation
Apr 29, 2025Abstract:While accurate and user-friendly Computer-Aided Design (CAD) is crucial for industrial design and manufacturing, existing methods still struggle to achieve this due to their over-simplified representations or architectures incapable of supporting multimodal design requirements. In this paper, we attempt to tackle this problem from both methods and datasets aspects. First, we propose a cascade MAR with topology predictor (CMT), the first multimodal framework for CAD generation based on Boundary Representation (B-Rep). Specifically, the cascade MAR can effectively capture the ``edge-counters-surface'' priors that are essential in B-Reps, while the topology predictor directly estimates topology in B-Reps from the compact tokens in MAR. Second, to facilitate large-scale training, we develop a large-scale multimodal CAD dataset, mmABC, which includes over 1.3 million B-Rep models with multimodal annotations, including point clouds, text descriptions, and multi-view images. Extensive experiments show the superior of CMT in both conditional and unconditional CAD generation tasks. For example, we improve Coverage and Valid ratio by +10.68% and +10.3%, respectively, compared to state-of-the-art methods on ABC in unconditional generation. CMT also improves +4.01 Chamfer on image conditioned CAD generation on mmABC. The dataset, code and pretrained network shall be released.
UniSTD: Towards Unified Spatio-Temporal Learning across Diverse Disciplines
Mar 26, 2025

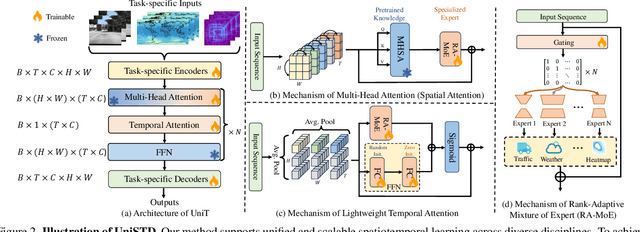

Abstract:Traditional spatiotemporal models generally rely on task-specific architectures, which limit their generalizability and scalability across diverse tasks due to domain-specific design requirements. In this paper, we introduce \textbf{UniSTD}, a unified Transformer-based framework for spatiotemporal modeling, which is inspired by advances in recent foundation models with the two-stage pretraining-then-adaption paradigm. Specifically, our work demonstrates that task-agnostic pretraining on 2D vision and vision-text datasets can build a generalizable model foundation for spatiotemporal learning, followed by specialized joint training on spatiotemporal datasets to enhance task-specific adaptability. To improve the learning capabilities across domains, our framework employs a rank-adaptive mixture-of-expert adaptation by using fractional interpolation to relax the discrete variables so that can be optimized in the continuous space. Additionally, we introduce a temporal module to incorporate temporal dynamics explicitly. We evaluate our approach on a large-scale dataset covering 10 tasks across 4 disciplines, demonstrating that a unified spatiotemporal model can achieve scalable, cross-task learning and support up to 10 tasks simultaneously within one model while reducing training costs in multi-domain applications. Code will be available at https://github.com/1hunters/UniSTD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge