Yuchen Cai
On Predictability of Reinforcement Learning Dynamics for Large Language Models
Oct 02, 2025Abstract:Recent advances in reasoning capabilities of large language models (LLMs) are largely driven by reinforcement learning (RL), yet the underlying parameter dynamics during RL training remain poorly understood. This work identifies two fundamental properties of RL-induced parameter updates in LLMs: (1) Rank-1 Dominance, where the top singular subspace of the parameter update matrix nearly fully determines reasoning improvements, recovering over 99\% of performance gains; and (2) Rank-1 Linear Dynamics, where this dominant subspace evolves linearly throughout training, enabling accurate prediction from early checkpoints. Extensive experiments across 8 LLMs and 7 algorithms validate the generalizability of these properties. More importantly, based on these findings, we propose AlphaRL, a plug-in acceleration framework that extrapolates the final parameter update using a short early training window, achieving up to 2.5 speedup while retaining \textgreater 96\% of reasoning performance without extra modules or hyperparameter tuning. This positions our finding as a versatile and practical tool for large-scale RL, opening a path toward principled, interpretable, and efficient training paradigm for LLMs.
DeltaEdit: Enhancing Sequential Editing in Large Language Models by Controlling Superimposed Noise
May 12, 2025Abstract:Sequential knowledge editing techniques aim to continuously update the knowledge in large language models at a low cost, preventing the models from generating outdated or incorrect information. However, existing sequential editing methods suffer from a significant decline in editing success rates after long-term editing. Through theoretical analysis and experiments, we identify that as the number of edits increases, the model's output increasingly deviates from the desired target, leading to a drop in editing success rates. We refer to this issue as the accumulation of superimposed noise problem. To address this, we identify the factors contributing to this deviation and propose DeltaEdit, a novel method that optimizes update parameters through a dynamic orthogonal constraints strategy, effectively reducing interference between edits to mitigate deviation. Experimental results demonstrate that DeltaEdit significantly outperforms existing methods in edit success rates and the retention of generalization capabilities, ensuring stable and reliable model performance even under extensive sequential editing.
COMET: Benchmark for Comprehensive Biological Multi-omics Evaluation Tasks and Language Models
Dec 13, 2024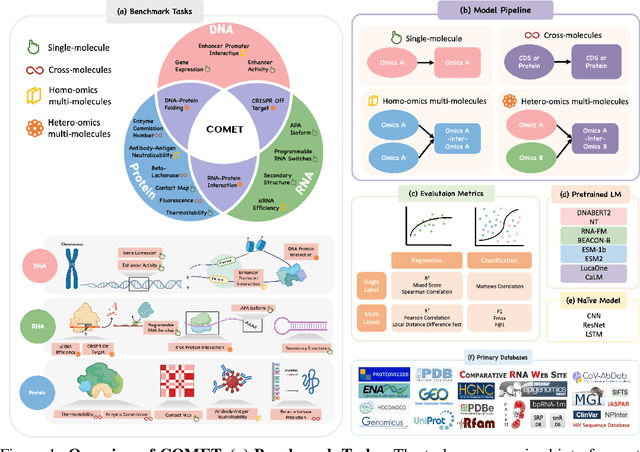
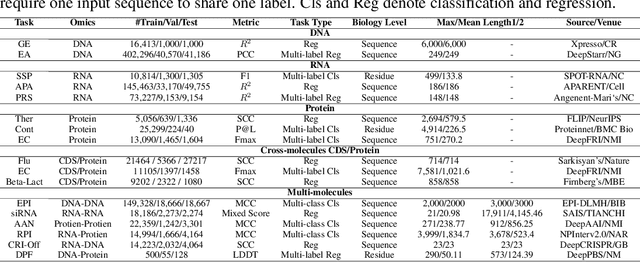
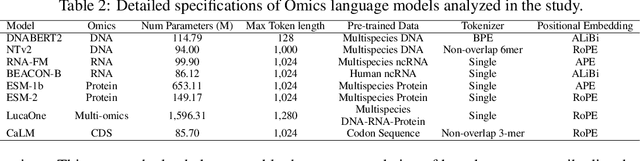
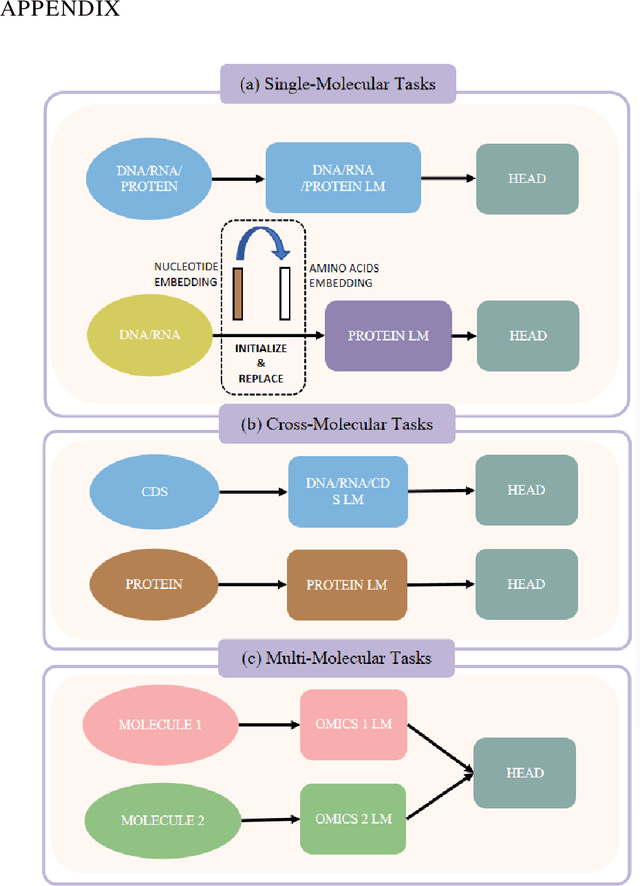
Abstract:As key elements within the central dogma, DNA, RNA, and proteins play crucial roles in maintaining life by guaranteeing accurate genetic expression and implementation. Although research on these molecules has profoundly impacted fields like medicine, agriculture, and industry, the diversity of machine learning approaches-from traditional statistical methods to deep learning models and large language models-poses challenges for researchers in choosing the most suitable models for specific tasks, especially for cross-omics and multi-omics tasks due to the lack of comprehensive benchmarks. To address this, we introduce the first comprehensive multi-omics benchmark COMET (Benchmark for Biological COmprehensive Multi-omics Evaluation Tasks and Language Models), designed to evaluate models across single-omics, cross-omics, and multi-omics tasks. First, we curate and develop a diverse collection of downstream tasks and datasets covering key structural and functional aspects in DNA, RNA, and proteins, including tasks that span multiple omics levels. Then, we evaluate existing foundational language models for DNA, RNA, and proteins, as well as the newly proposed multi-omics method, offering valuable insights into their performance in integrating and analyzing data from different biological modalities. This benchmark aims to define critical issues in multi-omics research and guide future directions, ultimately promoting advancements in understanding biological processes through integrated and different omics data analysis.
O-Edit: Orthogonal Subspace Editing for Language Model Sequential Editing
Oct 15, 2024Abstract:Large language models (LLMs) acquire knowledge during pre-training, but over time, this knowledge may become incorrect or outdated, necessitating updates after training. Knowledge editing techniques address this issue without the need for costly re-training. However, most existing methods are designed for single edits, and as the number of edits increases, they often cause a decline in the model's overall performance, posing significant challenges for sequential editing. To overcome this, we propose Orthogonal Subspace Editing, O-Edit. This algorithm orthogonalizes the direction of each knowledge update, minimizing interference between successive updates and reducing the impact of new updates on unrelated knowledge. Our approach does not require replaying previously edited data and processes each edit knowledge on time. It can perform thousands of edits on mainstream LLMs, achieving an average performance improvement that is 4.2 times better than existing methods while effectively preserving the model's performance on downstream tasks, all with minimal additional parameter overhead.
BEACON: Benchmark for Comprehensive RNA Tasks and Language Models
Jun 14, 2024Abstract:RNA plays a pivotal role in translating genetic instructions into functional outcomes, underscoring its importance in biological processes and disease mechanisms. Despite the emergence of numerous deep learning approaches for RNA, particularly universal RNA language models, there remains a significant lack of standardized benchmarks to assess the effectiveness of these methods. In this study, we introduce the first comprehensive RNA benchmark BEACON (\textbf{BE}nchm\textbf{A}rk for \textbf{CO}mprehensive R\textbf{N}A Task and Language Models). First, BEACON comprises 13 distinct tasks derived from extensive previous work covering structural analysis, functional studies, and engineering applications, enabling a comprehensive assessment of the performance of methods on various RNA understanding tasks. Second, we examine a range of models, including traditional approaches like CNNs, as well as advanced RNA foundation models based on language models, offering valuable insights into the task-specific performances of these models. Third, we investigate the vital RNA language model components from the tokenizer and positional encoding aspects. Notably, our findings emphasize the superiority of single nucleotide tokenization and the effectiveness of Attention with Linear Biases (ALiBi) over traditional positional encoding methods. Based on these insights, a simple yet strong baseline called BEACON-B is proposed, which can achieve outstanding performance with limited data and computational resources. The datasets and source code of our benchmark are available at https://github.com/terry-r123/RNABenchmark.
Editing Knowledge Representation of Language Lodel via Rephrased Prefix Prompts
Mar 21, 2024



Abstract:Neural language models (LMs) have been extensively trained on vast corpora to store factual knowledge about various aspects of the world described in texts. Current technologies typically employ knowledge editing methods or specific prompts to modify LM outputs. However, existing knowledge editing methods are costly and inefficient, struggling to produce appropriate text. Additionally, prompt engineering is opaque and requires significant effort to find suitable prompts. To address these issues, we introduce a new method called PSPEM (Prefix Soft Prompt Editing Method), that can be used for a lifetime with just one training. It resolves the inefficiencies and generalizability issues in knowledge editing methods and overcomes the opacity of prompt engineering by automatically seeking optimal soft prompts. Specifically, PSPEM utilizes a prompt encoder and an encoding converter to refine key information in prompts and uses prompt alignment techniques to guide model generation, ensuring text consistency and adherence to the intended structure and content, thereby maintaining an optimal balance between efficiency and accuracy. We have validated the effectiveness of PSPEM through knowledge editing and attribute inserting. On the COUNTERFACT dataset, PSPEM achieved nearly 100\% editing accuracy and demonstrated the highest level of fluency. We further analyzed the similarities between PSPEM and original prompts and their impact on the model's internals. The results indicate that PSPEM can serve as an alternative to original prompts, supporting the model in effective editing.
Locating and Mitigating Gender Bias in Large Language Models
Mar 21, 2024



Abstract:Large language models(LLM) are pre-trained on extensive corpora to learn facts and human cognition which contain human preferences. However, this process can inadvertently lead to these models acquiring biases and stereotypes prevalent in society. Prior research has typically tackled the issue of bias through a one-dimensional perspective, concentrating either on locating or mitigating it. This limited perspective has created obstacles in facilitating research on bias to synergistically complement and progressively build upon one another. In this study, we integrate the processes of locating and mitigating bias within a unified framework. Initially, we use causal mediation analysis to trace the causal effects of different components' activation within a large language model. Building on this, we propose the LSDM (Least Square Debias Method), a knowledge-editing based method for mitigating gender bias in occupational pronouns, and compare it against two baselines on three gender bias datasets and seven knowledge competency test datasets. The experimental results indicate that the primary contributors to gender bias are the bottom MLP modules acting on the last token of occupational pronouns and the top attention module acting on the final word in the sentence. Furthermore, LSDM mitigates gender bias in the model more effectively than the other baselines, while fully preserving the model's capabilities in all other aspects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge