Siqi Sun
InternAgent-1.5: A Unified Agentic Framework for Long-Horizon Autonomous Scientific Discovery
Feb 09, 2026Abstract:We introduce InternAgent-1.5, a unified system designed for end-to-end scientific discovery across computational and empirical domains. The system is built on a structured architecture composed of three coordinated subsystems for generation, verification, and evolution. These subsystems are supported by foundational capabilities for deep research, solution optimization, and long horizon memory. The architecture allows InternAgent-1.5 to operate continuously across extended discovery cycles while maintaining coherent and improving behavior. It also enables the system to coordinate computational modeling and laboratory experimentation within a single unified system. We evaluate InternAgent-1.5 on scientific reasoning benchmarks such as GAIA, HLE, GPQA, and FrontierScience, and the system achieves leading performance that demonstrates strong foundational capabilities. Beyond these benchmarks, we further assess two categories of discovery tasks. In algorithm discovery tasks, InternAgent-1.5 autonomously designs competitive methods for core machine learning problems. In empirical discovery tasks, it executes complete computational or wet lab experiments and produces scientific findings in earth, life, biological, and physical domains. Overall, these results show that InternAgent-1.5 provides a general and scalable framework for autonomous scientific discovery.
Reflection Pretraining Enables Token-Level Self-Correction in Biological Sequence Models
Dec 24, 2025



Abstract:Chain-of-Thought (CoT) prompting has significantly advanced task-solving capabilities in natural language processing with large language models. Unlike standard prompting, CoT encourages the model to generate intermediate reasoning steps, non-answer tokens, that help guide the model toward more accurate final outputs. These intermediate steps enable more complex reasoning processes such as error correction, memory management, future planning, and self-reflection. However, applying CoT to non-natural language domains, such as protein and RNA language models, is not yet possible, primarily due to the limited expressiveness of their token spaces (e.g., amino acid tokens). In this work, we propose and define the concept of language expressiveness: the ability of a given language, using its tokens and grammar, to encode information. We show that the limited expressiveness of protein language severely restricts the applicability of CoT-style reasoning. To overcome this, we introduce reflection pretraining, for the first time in a biological sequence model, which enables the model to engage in intermediate reasoning through the generation of auxiliary "thinking tokens" beyond simple answer tokens. Theoretically, we demonstrate that our augmented token set significantly enhances biological language expressiveness, thereby improving the overall reasoning capacity of the model. Experimentally, our pretraining approach teaches protein models to self-correct and leads to substantial performance gains compared to standard pretraining.
Probing Scientific General Intelligence of LLMs with Scientist-Aligned Workflows
Dec 18, 2025Abstract:Despite advances in scientific AI, a coherent framework for Scientific General Intelligence (SGI)-the ability to autonomously conceive, investigate, and reason across scientific domains-remains lacking. We present an operational SGI definition grounded in the Practical Inquiry Model (PIM: Deliberation, Conception, Action, Perception) and operationalize it via four scientist-aligned tasks: deep research, idea generation, dry/wet experiments, and experimental reasoning. SGI-Bench comprises over 1,000 expert-curated, cross-disciplinary samples inspired by Science's 125 Big Questions, enabling systematic evaluation of state-of-the-art LLMs. Results reveal gaps: low exact match (10--20%) in deep research despite step-level alignment; ideas lacking feasibility and detail; high code executability but low execution result accuracy in dry experiments; low sequence fidelity in wet protocols; and persistent multimodal comparative-reasoning challenges. We further introduce Test-Time Reinforcement Learning (TTRL), which optimizes retrieval-augmented novelty rewards at inference, enhancing hypothesis novelty without reference answer. Together, our PIM-grounded definition, workflow-centric benchmark, and empirical insights establish a foundation for AI systems that genuinely participate in scientific discovery.
Accurate de novo sequencing of the modified proteome with OmniNovo
Dec 13, 2025Abstract:Post-translational modifications (PTMs) serve as a dynamic chemical language regulating protein function, yet current proteomic methods remain blind to a vast portion of the modified proteome. Standard database search algorithms suffer from a combinatorial explosion of search spaces, limiting the identification of uncharacterized or complex modifications. Here we introduce OmniNovo, a unified deep learning framework for reference-free sequencing of unmodified and modified peptides directly from tandem mass spectra. Unlike existing tools restricted to specific modification types, OmniNovo learns universal fragmentation rules to decipher diverse PTMs within a single coherent model. By integrating a mass-constrained decoding algorithm with rigorous false discovery rate estimation, OmniNovo achieves state-of-the-art accuracy, identifying 51\% more peptides than standard approaches at a 1\% false discovery rate. Crucially, the model generalizes to biological sites unseen during training, illuminating the dark matter of the proteome and enabling unbiased comprehensive analysis of cellular regulation.
ATLAS: A High-Difficulty, Multidisciplinary Benchmark for Frontier Scientific Reasoning
Nov 18, 2025Abstract:The rapid advancement of Large Language Models (LLMs) has led to performance saturation on many established benchmarks, questioning their ability to distinguish frontier models. Concurrently, existing high-difficulty benchmarks often suffer from narrow disciplinary focus, oversimplified answer formats, and vulnerability to data contamination, creating a fidelity gap with real-world scientific inquiry. To address these challenges, we introduce ATLAS (AGI-Oriented Testbed for Logical Application in Science), a large-scale, high-difficulty, and cross-disciplinary evaluation suite composed of approximately 800 original problems. Developed by domain experts (PhD-level and above), ATLAS spans seven core scientific fields: mathematics, physics, chemistry, biology, computer science, earth science, and materials science. Its key features include: (1) High Originality and Contamination Resistance, with all questions newly created or substantially adapted to prevent test data leakage; (2) Cross-Disciplinary Focus, designed to assess models' ability to integrate knowledge and reason across scientific domains; (3) High-Fidelity Answers, prioritizing complex, open-ended answers involving multi-step reasoning and LaTeX-formatted expressions over simple multiple-choice questions; and (4) Rigorous Quality Control, employing a multi-stage process of expert peer review and adversarial testing to ensure question difficulty, scientific value, and correctness. We also propose a robust evaluation paradigm using a panel of LLM judges for automated, nuanced assessment of complex answers. Preliminary results on leading models demonstrate ATLAS's effectiveness in differentiating their advanced scientific reasoning capabilities. We plan to develop ATLAS into a long-term, open, community-driven platform to provide a reliable "ruler" for progress toward Artificial General Intelligence.
TimeSeriesScientist: A General-Purpose AI Agent for Time Series Analysis
Oct 02, 2025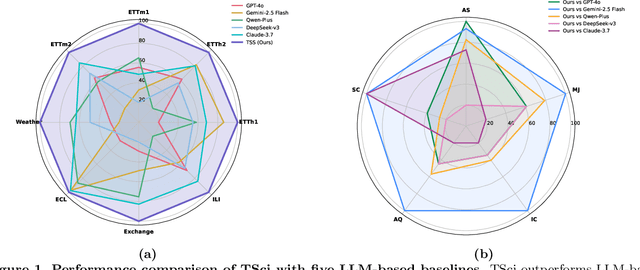
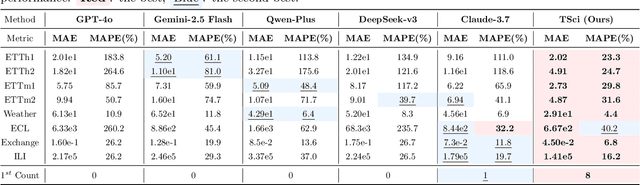
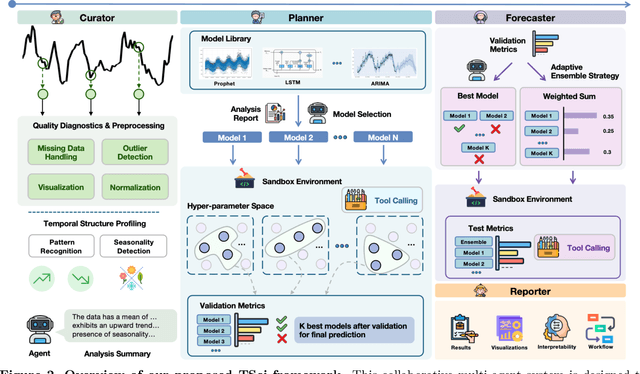
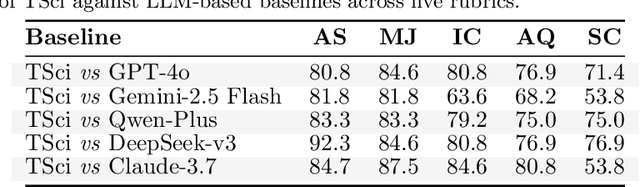
Abstract:Time series forecasting is central to decision-making in domains as diverse as energy, finance, climate, and public health. In practice, forecasters face thousands of short, noisy series that vary in frequency, quality, and horizon, where the dominant cost lies not in model fitting, but in the labor-intensive preprocessing, validation, and ensembling required to obtain reliable predictions. Prevailing statistical and deep learning models are tailored to specific datasets or domains and generalize poorly. A general, domain-agnostic framework that minimizes human intervention is urgently in demand. In this paper, we introduce TimeSeriesScientist (TSci), the first LLM-driven agentic framework for general time series forecasting. The framework comprises four specialized agents: Curator performs LLM-guided diagnostics augmented by external tools that reason over data statistics to choose targeted preprocessing; Planner narrows the hypothesis space of model choice by leveraging multi-modal diagnostics and self-planning over the input; Forecaster performs model fitting and validation and, based on the results, adaptively selects the best model configuration as well as ensemble strategy to make final predictions; and Reporter synthesizes the whole process into a comprehensive, transparent report. With transparent natural-language rationales and comprehensive reports, TSci transforms the forecasting workflow into a white-box system that is both interpretable and extensible across tasks. Empirical results on eight established benchmarks demonstrate that TSci consistently outperforms both statistical and LLM-based baselines, reducing forecast error by an average of 10.4% and 38.2%, respectively. Moreover, TSci produces a clear and rigorous report that makes the forecasting workflow more transparent and interpretable.
Curriculum Learning for Biological Sequence Prediction: The Case of De Novo Peptide Sequencing
Jun 16, 2025Abstract:Peptide sequencing-the process of identifying amino acid sequences from mass spectrometry data-is a fundamental task in proteomics. Non-Autoregressive Transformers (NATs) have proven highly effective for this task, outperforming traditional methods. Unlike autoregressive models, which generate tokens sequentially, NATs predict all positions simultaneously, leveraging bidirectional context through unmasked self-attention. However, existing NAT approaches often rely on Connectionist Temporal Classification (CTC) loss, which presents significant optimization challenges due to CTC's complexity and increases the risk of training failures. To address these issues, we propose an improved non-autoregressive peptide sequencing model that incorporates a structured protein sequence curriculum learning strategy. This approach adjusts protein's learning difficulty based on the model's estimated protein generational capabilities through a sampling process, progressively learning peptide generation from simple to complex sequences. Additionally, we introduce a self-refining inference-time module that iteratively enhances predictions using learned NAT token embeddings, improving sequence accuracy at a fine-grained level. Our curriculum learning strategy reduces NAT training failures frequency by more than 90% based on sampled training over various data distributions. Evaluations on nine benchmark species demonstrate that our approach outperforms all previous methods across multiple metrics and species.
Scientists' First Exam: Probing Cognitive Abilities of MLLM via Perception, Understanding, and Reasoning
Jun 12, 2025Abstract:Scientific discoveries increasingly rely on complex multimodal reasoning based on information-intensive scientific data and domain-specific expertise. Empowered by expert-level scientific benchmarks, scientific Multimodal Large Language Models (MLLMs) hold the potential to significantly enhance this discovery process in realistic workflows. However, current scientific benchmarks mostly focus on evaluating the knowledge understanding capabilities of MLLMs, leading to an inadequate assessment of their perception and reasoning abilities. To address this gap, we present the Scientists' First Exam (SFE) benchmark, designed to evaluate the scientific cognitive capacities of MLLMs through three interconnected levels: scientific signal perception, scientific attribute understanding, scientific comparative reasoning. Specifically, SFE comprises 830 expert-verified VQA pairs across three question types, spanning 66 multimodal tasks across five high-value disciplines. Extensive experiments reveal that current state-of-the-art GPT-o3 and InternVL-3 achieve only 34.08% and 26.52% on SFE, highlighting significant room for MLLMs to improve in scientific realms. We hope the insights obtained in SFE will facilitate further developments in AI-enhanced scientific discoveries.
Universal Biological Sequence Reranking for Improved De Novo Peptide Sequencing
May 23, 2025Abstract:De novo peptide sequencing is a critical task in proteomics. However, the performance of current deep learning-based methods is limited by the inherent complexity of mass spectrometry data and the heterogeneous distribution of noise signals, leading to data-specific biases. We present RankNovo, the first deep reranking framework that enhances de novo peptide sequencing by leveraging the complementary strengths of multiple sequencing models. RankNovo employs a list-wise reranking approach, modeling candidate peptides as multiple sequence alignments and utilizing axial attention to extract informative features across candidates. Additionally, we introduce two new metrics, PMD (Peptide Mass Deviation) and RMD (residual Mass Deviation), which offer delicate supervision by quantifying mass differences between peptides at both the sequence and residue levels. Extensive experiments demonstrate that RankNovo not only surpasses its base models used to generate training candidates for reranking pre-training, but also sets a new state-of-the-art benchmark. Moreover, RankNovo exhibits strong zero-shot generalization to unseen models whose generations were not exposed during training, highlighting its robustness and potential as a universal reranking framework for peptide sequencing. Our work presents a novel reranking strategy that fundamentally challenges existing single-model paradigms and advances the frontier of accurate de novo sequencing. Our source code is provided on GitHub.
AlignRAG: An Adaptable Framework for Resolving Misalignments in Retrieval-Aware Reasoning of RAG
Apr 21, 2025Abstract:Retrieval-augmented generation (RAG) has emerged as a foundational paradigm for knowledge-grounded text generation. However, existing RAG pipelines often fail to ensure that the reasoning trajectories align with the evidential constraints imposed by retrieved content. In this paper, we reframe RAG as a problem of retrieval-aware reasoning and identify a core challenge: reasoning misalignment-the mismatch between a model's reasoning trajectory and the retrieved evidence. To address this challenge, we propose AlignRAG, a novel test-time framework that mitigates reasoning misalignment through iterative Critique-Driven Alignment (CDA) steps. In contrast to prior approaches that rely on static training or post-hoc selection, AlignRAG actively refines reasoning trajectories during inference by enforcing fine-grained alignment with evidence. Our framework introduces a new paradigm for retrieval-aware reasoning by: (1) constructing context-rich training corpora; (2) generating contrastive critiques from preference-aware reasoning trajectories; (3) training a dedicated \textit{Critic Language Model (CLM)} to identify reasoning misalignments; and (4) applying CDA steps to optimize reasoning trajectories iteratively. Empirical results demonstrate that AlignRAG consistently outperforms all baselines and could integrate as a plug-and-play module into existing RAG pipelines without further changes. By reconceptualizing RAG as a structured reasoning trajectory and establishing the test-time framework for correcting reasoning misalignments in RAG, AlignRAG provides practical advancements for retrieval-aware generation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge