Qianyuan Zhang
How Order-Sensitive Are LLMs? OrderProbe for Deterministic Structural Reconstruction
Jan 13, 2026Abstract:Large language models (LLMs) excel at semantic understanding, yet their ability to reconstruct internal structure from scrambled inputs remains underexplored. Sentence-level restoration is ill-posed for automated evaluation because multiple valid word orders often exist. We introduce OrderProbe, a deterministic benchmark for structural reconstruction using fixed four-character expressions in Chinese, Japanese, and Korean, which have a unique canonical order and thus support exact-match scoring. We further propose a diagnostic framework that evaluates models beyond recovery accuracy, including semantic fidelity, logical validity, consistency, robustness sensitivity, and information density. Experiments on twelve widely used LLMs show that structural reconstruction remains difficult even for frontier systems: zero-shot recovery frequently falls below 35%. We also observe a consistent dissociation between semantic recall and structural planning, suggesting that structural robustness is not an automatic byproduct of semantic competence.
COMET: Benchmark for Comprehensive Biological Multi-omics Evaluation Tasks and Language Models
Dec 13, 2024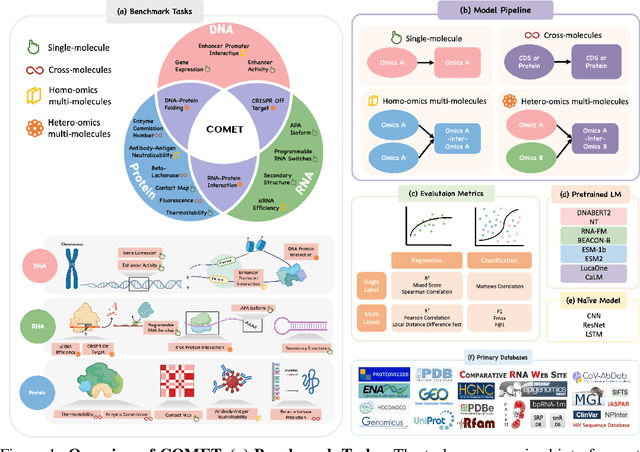
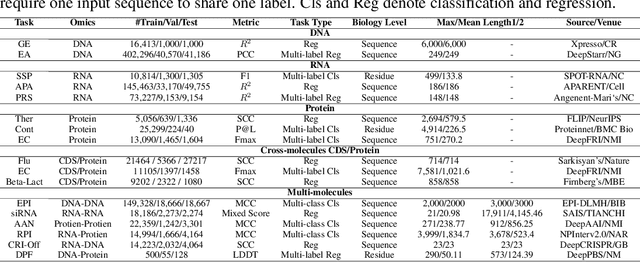
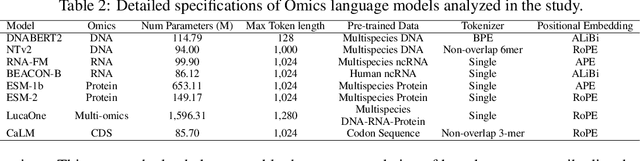
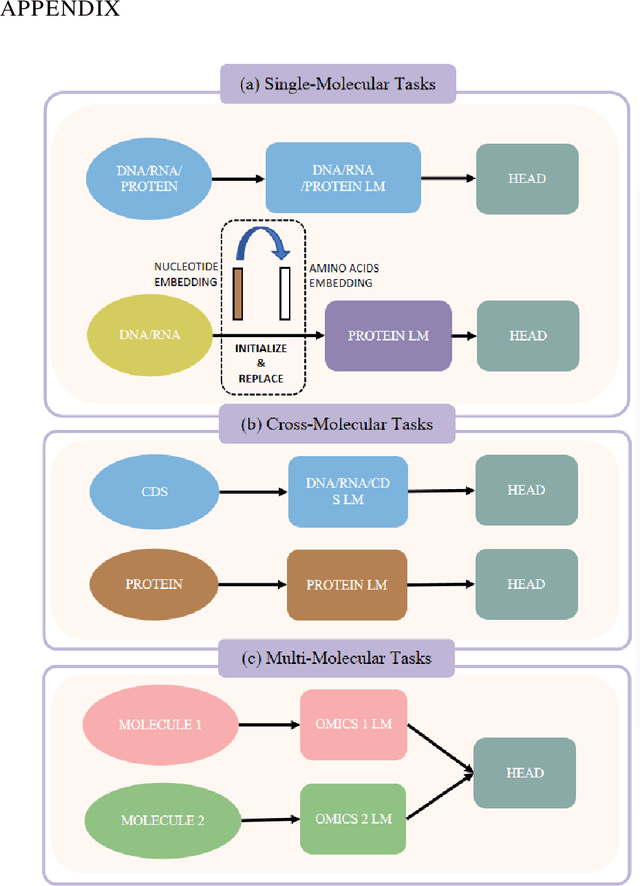
Abstract:As key elements within the central dogma, DNA, RNA, and proteins play crucial roles in maintaining life by guaranteeing accurate genetic expression and implementation. Although research on these molecules has profoundly impacted fields like medicine, agriculture, and industry, the diversity of machine learning approaches-from traditional statistical methods to deep learning models and large language models-poses challenges for researchers in choosing the most suitable models for specific tasks, especially for cross-omics and multi-omics tasks due to the lack of comprehensive benchmarks. To address this, we introduce the first comprehensive multi-omics benchmark COMET (Benchmark for Biological COmprehensive Multi-omics Evaluation Tasks and Language Models), designed to evaluate models across single-omics, cross-omics, and multi-omics tasks. First, we curate and develop a diverse collection of downstream tasks and datasets covering key structural and functional aspects in DNA, RNA, and proteins, including tasks that span multiple omics levels. Then, we evaluate existing foundational language models for DNA, RNA, and proteins, as well as the newly proposed multi-omics method, offering valuable insights into their performance in integrating and analyzing data from different biological modalities. This benchmark aims to define critical issues in multi-omics research and guide future directions, ultimately promoting advancements in understanding biological processes through integrated and different omics data analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge