Dong Yuan
Optimal Look-back Horizon for Time Series Forecasting in Federated Learning
Nov 18, 2025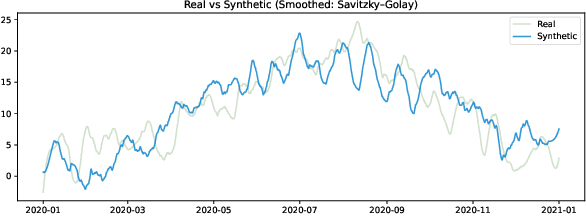
Abstract:Selecting an appropriate look-back horizon remains a fundamental challenge in time series forecasting (TSF), particularly in the federated learning scenarios where data is decentralized, heterogeneous, and often non-independent. While recent work has explored horizon selection by preserving forecasting-relevant information in an intrinsic space, these approaches are primarily restricted to centralized and independently distributed settings. This paper presents a principled framework for adaptive horizon selection in federated time series forecasting through an intrinsic space formulation. We introduce a synthetic data generator (SDG) that captures essential temporal structures in client data, including autoregressive dependencies, seasonality, and trend, while incorporating client-specific heterogeneity. Building on this model, we define a transformation that maps time series windows into an intrinsic representation space with well-defined geometric and statistical properties. We then derive a decomposition of the forecasting loss into a Bayesian term, which reflects irreducible uncertainty, and an approximation term, which accounts for finite-sample effects and limited model capacity. Our analysis shows that while increasing the look-back horizon improves the identifiability of deterministic patterns, it also increases approximation error due to higher model complexity and reduced sample efficiency. We prove that the total forecasting loss is minimized at the smallest horizon where the irreducible loss starts to saturate, while the approximation loss continues to rise. This work provides a rigorous theoretical foundation for adaptive horizon selection for time series forecasting in federated learning.
Scaling Law Analysis in Federated Learning: How to Select the Optimal Model Size?
Nov 15, 2025Abstract:The recent success of large language models (LLMs) has sparked a growing interest in training large-scale models. As the model size continues to scale, concerns are growing about the depletion of high-quality, well-curated training data. This has led practitioners to explore training approaches like Federated Learning (FL), which can leverage the abundant data on edge devices while maintaining privacy. However, the decentralization of training datasets in FL introduces challenges to scaling large models, a topic that remains under-explored. This paper fills this gap and provides qualitative insights on generalizing the previous model scaling experience to federated learning scenarios. Specifically, we derive a PAC-Bayes (Probably Approximately Correct Bayesian) upper bound for the generalization error of models trained with stochastic algorithms in federated settings and quantify the impact of distributed training data on the optimal model size by finding the analytic solution of model size that minimizes this bound. Our theoretical results demonstrate that the optimal model size has a negative power law relationship with the number of clients if the total training compute is unchanged. Besides, we also find that switching to FL with the same training compute will inevitably reduce the upper bound of generalization performance that the model can achieve through training, and that estimating the optimal model size in federated scenarios should depend on the average training compute across clients. Furthermore, we also empirically validate the correctness of our results with extensive training runs on different models, network settings, and datasets.
GuardFed: A Trustworthy Federated Learning Framework Against Dual-Facet Attacks
Nov 12, 2025Abstract:Federated learning (FL) enables privacy-preserving collaborative model training but remains vulnerable to adversarial behaviors that compromise model utility or fairness across sensitive groups. While extensive studies have examined attacks targeting either objective, strategies that simultaneously degrade both utility and fairness remain largely unexplored. To bridge this gap, we introduce the Dual-Facet Attack (DFA), a novel threat model that concurrently undermines predictive accuracy and group fairness. Two variants, Synchronous DFA (S-DFA) and Split DFA (Sp-DFA), are further proposed to capture distinct real-world collusion scenarios. Experimental results show that existing robust FL defenses, including hybrid aggregation schemes, fail to resist DFAs effectively. To counter these threats, we propose GuardFed, a self-adaptive defense framework that maintains a fairness-aware reference model using a small amount of clean server data augmented with synthetic samples. In each training round, GuardFed computes a dual-perspective trust score for every client by jointly evaluating its utility deviation and fairness degradation, thereby enabling selective aggregation of trustworthy updates. Extensive experiments on real-world datasets demonstrate that GuardFed consistently preserves both accuracy and fairness under diverse non-IID and adversarial conditions, achieving state-of-the-art performance compared with existing robust FL methods.
PUMPS: Skeleton-Agnostic Point-based Universal Motion Pre-Training for Synthesis in Human Motion Tasks
Jul 27, 2025Abstract:Motion skeletons drive 3D character animation by transforming bone hierarchies, but differences in proportions or structure make motion data hard to transfer across skeletons, posing challenges for data-driven motion synthesis. Temporal Point Clouds (TPCs) offer an unstructured, cross-compatible motion representation. Though reversible with skeletons, TPCs mainly serve for compatibility, not for direct motion task learning. Doing so would require data synthesis capabilities for the TPC format, which presents unexplored challenges regarding its unique temporal consistency and point identifiability. Therefore, we propose PUMPS, the primordial autoencoder architecture for TPC data. PUMPS independently reduces frame-wise point clouds into sampleable feature vectors, from which a decoder extracts distinct temporal points using latent Gaussian noise vectors as sampling identifiers. We introduce linear assignment-based point pairing to optimise the TPC reconstruction process, and negate the use of expensive point-wise attention mechanisms in the architecture. Using these latent features, we pre-train a motion synthesis model capable of performing motion prediction, transition generation, and keyframe interpolation. For these pre-training tasks, PUMPS performs remarkably well even without native dataset supervision, matching state-of-the-art performance. When fine-tuned for motion denoising or estimation, PUMPS outperforms many respective methods without deviating from its generalist architecture.
Evaluate Summarization in Fine-Granularity: Auto Evaluation with LLM
Dec 27, 2024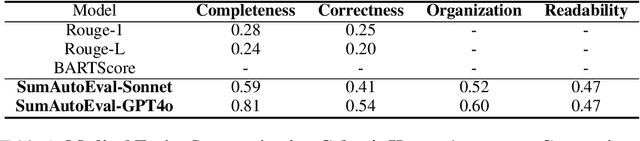
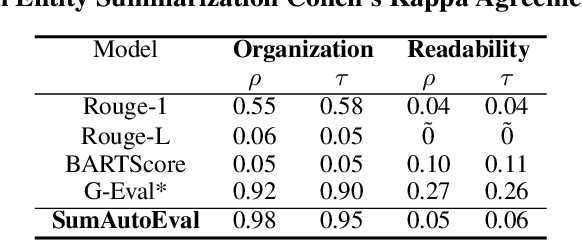
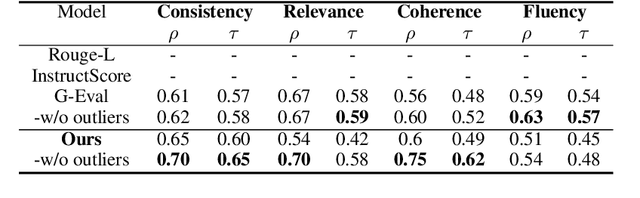
Abstract:Due to the exponential growth of information and the need for efficient information consumption the task of summarization has gained paramount importance. Evaluating summarization accurately and objectively presents significant challenges, particularly when dealing with long and unstructured texts rich in content. Existing methods, such as ROUGE (Lin, 2004) and embedding similarities, often yield scores that have low correlation with human judgements and are also not intuitively understandable, making it difficult to gauge the true quality of the summaries. LLMs can mimic human in giving subjective reviews but subjective scores are hard to interpret and justify. They can be easily manipulated by altering the models and the tones of the prompts. In this paper, we introduce a novel evaluation methodology and tooling designed to address these challenges, providing a more comprehensive, accurate and interpretable assessment of summarization outputs. Our method (SumAutoEval) proposes and evaluates metrics at varying granularity levels, giving objective scores on 4 key dimensions such as completeness, correctness, Alignment and readability. We empirically demonstrate, that SumAutoEval enhances the understanding of output quality with better human correlation.
Biology Instructions: A Dataset and Benchmark for Multi-Omics Sequence Understanding Capability of Large Language Models
Dec 26, 2024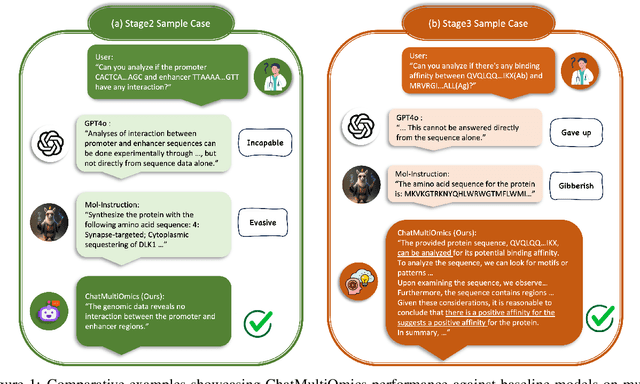
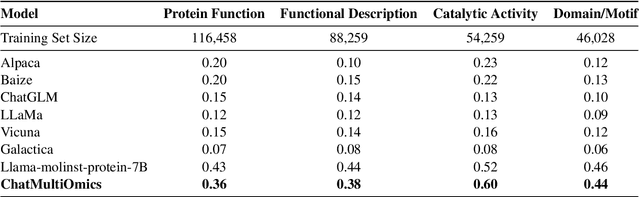
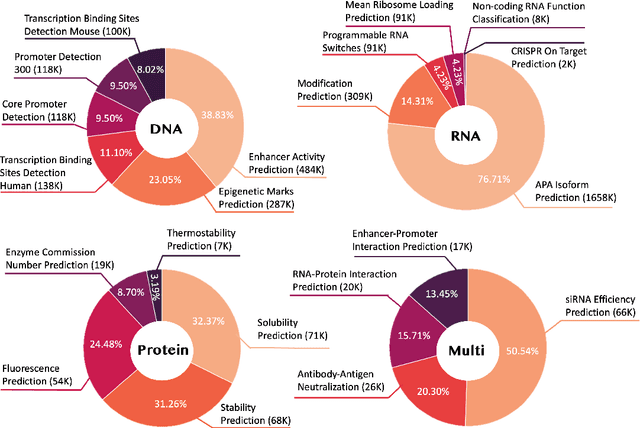
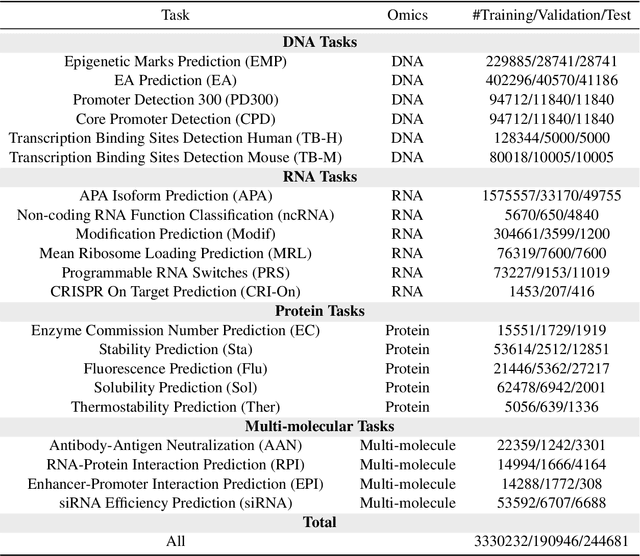
Abstract:Large language models have already demonstrated their formidable capabilities in general domains, ushering in a revolutionary transformation. However, exploring and exploiting the extensive knowledge of these models to comprehend multi-omics biology remains underexplored. To fill this research gap, we first introduce Biology-Instructions, the first large-scale multi-omics biological sequences-related instruction-tuning dataset including DNA, RNA, proteins, and multi-molecules, designed to bridge the gap between large language models (LLMs) and complex biological sequences-related tasks. This dataset can enhance the versatility of LLMs by integrating diverse biological sequenced-based prediction tasks with advanced reasoning capabilities, while maintaining conversational fluency. Additionally, we reveal significant performance limitations in even state-of-the-art LLMs on biological sequence-related multi-omics tasks without specialized pre-training and instruction-tuning. We further develop a strong baseline called ChatMultiOmics with a novel three-stage training pipeline, demonstrating the powerful ability to understand biology by using Biology-Instructions. Biology-Instructions and ChatMultiOmics are publicly available and crucial resources for enabling more effective integration of LLMs with multi-omics sequence analysis.
COMET: Benchmark for Comprehensive Biological Multi-omics Evaluation Tasks and Language Models
Dec 13, 2024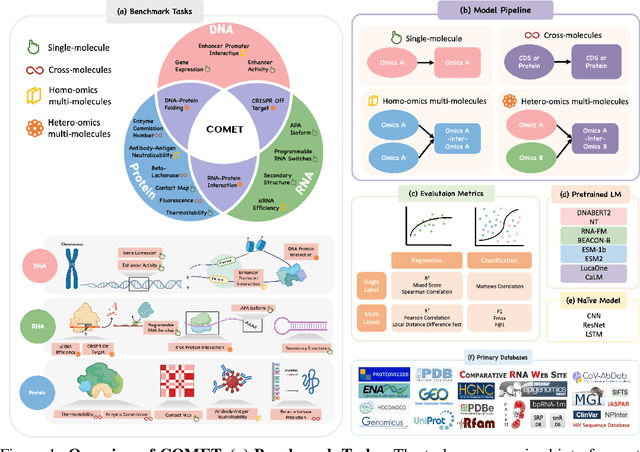
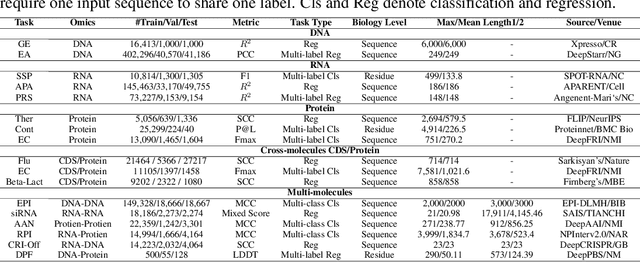
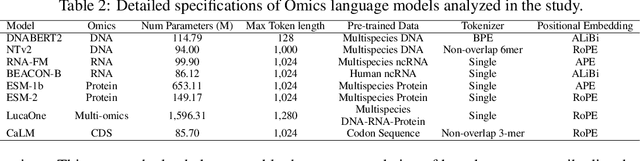
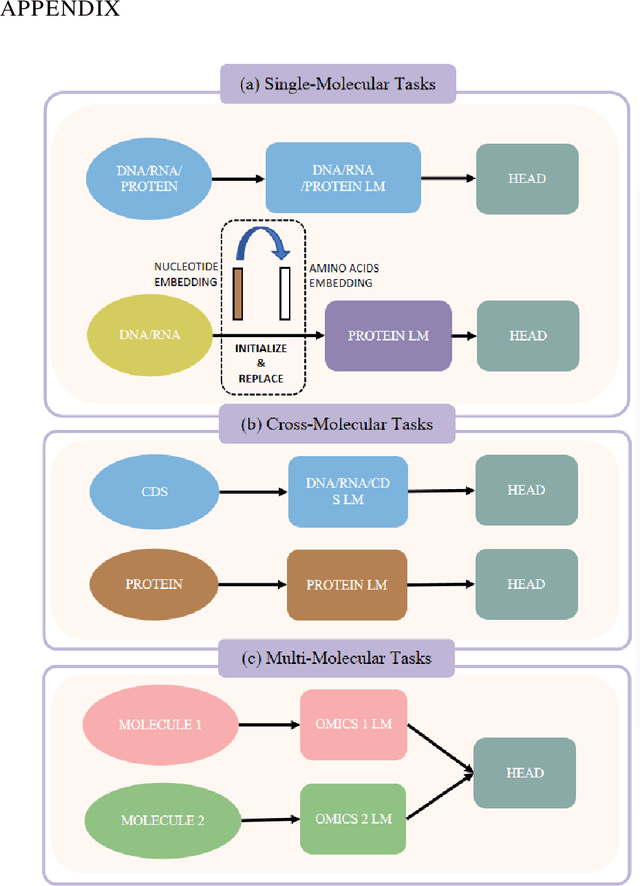
Abstract:As key elements within the central dogma, DNA, RNA, and proteins play crucial roles in maintaining life by guaranteeing accurate genetic expression and implementation. Although research on these molecules has profoundly impacted fields like medicine, agriculture, and industry, the diversity of machine learning approaches-from traditional statistical methods to deep learning models and large language models-poses challenges for researchers in choosing the most suitable models for specific tasks, especially for cross-omics and multi-omics tasks due to the lack of comprehensive benchmarks. To address this, we introduce the first comprehensive multi-omics benchmark COMET (Benchmark for Biological COmprehensive Multi-omics Evaluation Tasks and Language Models), designed to evaluate models across single-omics, cross-omics, and multi-omics tasks. First, we curate and develop a diverse collection of downstream tasks and datasets covering key structural and functional aspects in DNA, RNA, and proteins, including tasks that span multiple omics levels. Then, we evaluate existing foundational language models for DNA, RNA, and proteins, as well as the newly proposed multi-omics method, offering valuable insights into their performance in integrating and analyzing data from different biological modalities. This benchmark aims to define critical issues in multi-omics research and guide future directions, ultimately promoting advancements in understanding biological processes through integrated and different omics data analysis.
CAKD: A Correlation-Aware Knowledge Distillation Framework Based on Decoupling Kullback-Leibler Divergence
Oct 17, 2024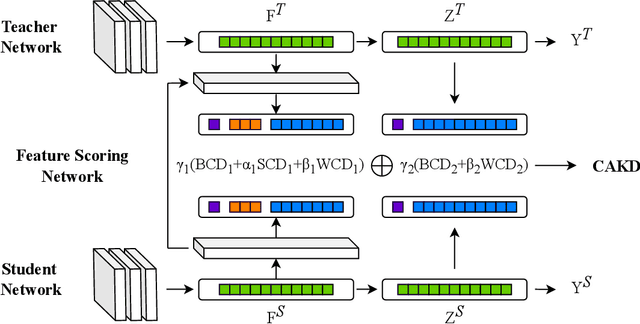
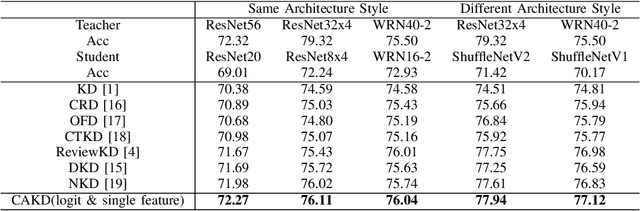
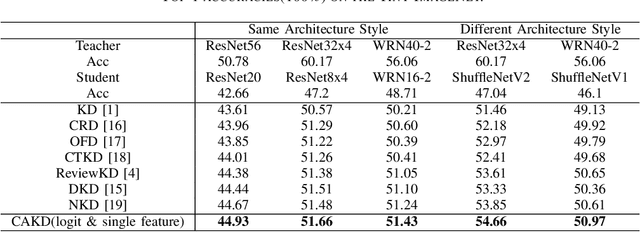
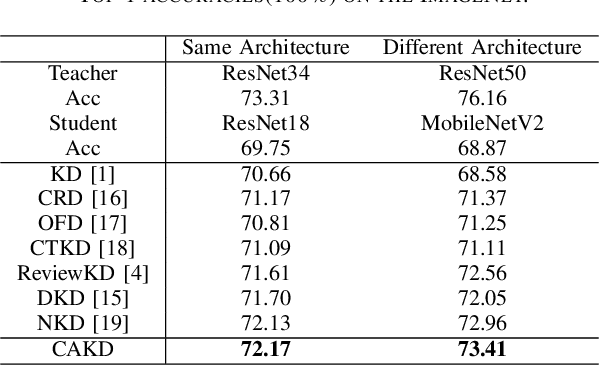
Abstract:In knowledge distillation, a primary focus has been on transforming and balancing multiple distillation components. In this work, we emphasize the importance of thoroughly examining each distillation component, as we observe that not all elements are equally crucial. From this perspective,we decouple the Kullback-Leibler (KL) divergence into three unique elements: Binary Classification Divergence (BCD), Strong Correlation Divergence (SCD), and Weak Correlation Divergence (WCD). Each of these elements presents varying degrees of influence. Leveraging these insights, we present the Correlation-Aware Knowledge Distillation (CAKD) framework. CAKD is designed to prioritize the facets of the distillation components that have the most substantial influence on predictions, thereby optimizing knowledge transfer from teacher to student models. Our experiments demonstrate that adjusting the effect of each element enhances the effectiveness of knowledge transformation. Furthermore, evidence shows that our novel CAKD framework consistently outperforms the baseline across diverse models and datasets. Our work further highlights the importance and effectiveness of closely examining the impact of different parts of distillation process.
Temporal Attention for Cross-View Sequential Image Localization
Aug 28, 2024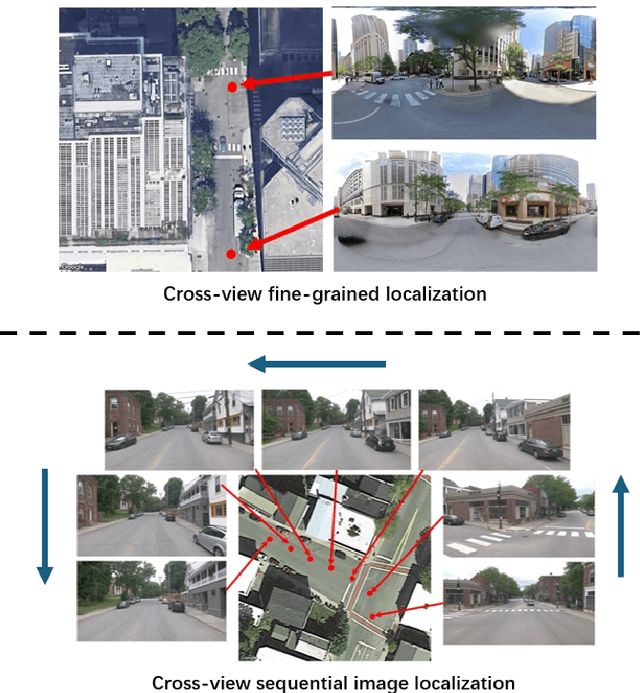
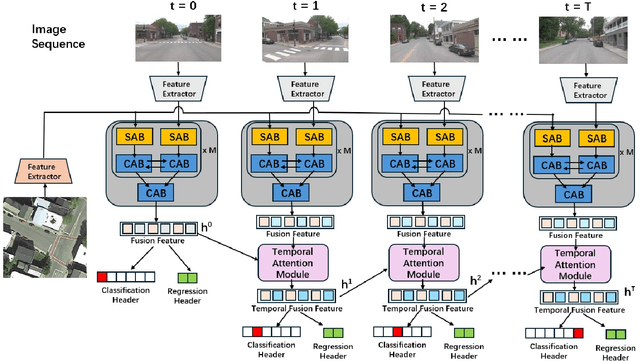
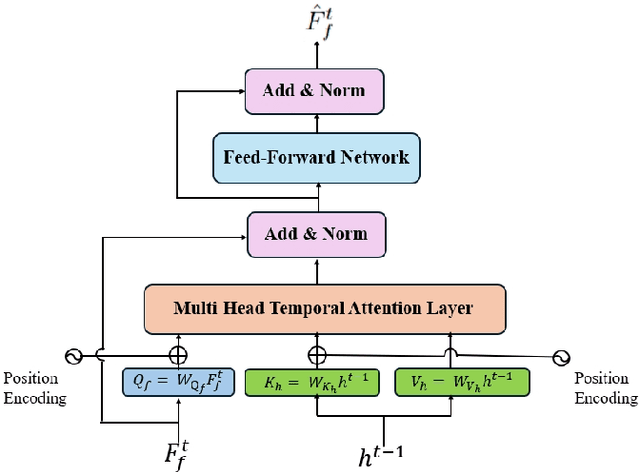

Abstract:This paper introduces a novel approach to enhancing cross-view localization, focusing on the fine-grained, sequential localization of street-view images within a single known satellite image patch, a significant departure from traditional one-to-one image retrieval methods. By expanding to sequential image fine-grained localization, our model, equipped with a novel Temporal Attention Module (TAM), leverages contextual information to significantly improve sequential image localization accuracy. Our method shows substantial reductions in both mean and median localization errors on the Cross-View Image Sequence (CVIS) dataset, outperforming current state-of-the-art single-image localization techniques. Additionally, by adapting the KITTI-CVL dataset into sequential image sets, we not only offer a more realistic dataset for future research but also demonstrate our model's robust generalization capabilities across varying times and areas, evidenced by a 75.3% reduction in mean distance error in cross-view sequential image localization.
Radio Frequency Signal based Human Silhouette Segmentation: A Sequential Diffusion Approach
Jul 27, 2024



Abstract:Radio frequency (RF) signals have been proved to be flexible for human silhouette segmentation (HSS) under complex environments. Existing studies are mainly based on a one-shot approach, which lacks a coherent projection ability from the RF domain. Additionally, the spatio-temporal patterns have not been fully explored for human motion dynamics in HSS. Therefore, we propose a two-stage Sequential Diffusion Model (SDM) to progressively synthesize high-quality segmentation jointly with the considerations on motion dynamics. Cross-view transformation blocks are devised to guide the diffusion model in a multi-scale manner for comprehensively characterizing human related patterns in an individual frame such as directional projection from signal planes. Moreover, spatio-temporal blocks are devised to fine-tune the frame-level model to incorporate spatio-temporal contexts and motion dynamics, enhancing the consistency of the segmentation maps. Comprehensive experiments on a public benchmark -- HIBER demonstrate the state-of-the-art performance of our method with an IoU 0.732. Our code is available at https://github.com/ph-w2000/SDM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge