Dongzhan Zhou
Discovery of Interpretable Physical Laws in Materials via Language-Model-Guided Symbolic Regression
Feb 26, 2026Abstract:Discovering interpretable physical laws from high-dimensional data is a fundamental challenge in scientific research. Traditional methods, such as symbolic regression, often produce complex, unphysical formulas when searching a vast space of possible forms. We introduce a framework that guides the search process by leveraging the embedded scientific knowledge of large language models, enabling efficient identification of physical laws in the data. We validate our approach by modeling key properties of perovskite materials. Our method mitigates the combinatorial explosion commonly encountered in traditional symbolic regression, reducing the effective search space by a factor of approximately $10^5$. A set of novel formulas for bulk modulus, band gap, and oxygen evolution reaction activity are identified, which not only provide meaningful physical insights but also outperform previous formulas in accuracy and simplicity.
AnyTouch 2: General Optical Tactile Representation Learning For Dynamic Tactile Perception
Feb 10, 2026Abstract:Real-world contact-rich manipulation demands robots to perceive temporal tactile feedback, capture subtle surface deformations, and reason about object properties as well as force dynamics. Although optical tactile sensors are uniquely capable of providing such rich information, existing tactile datasets and models remain limited. These resources primarily focus on object-level attributes (e.g., material) while largely overlooking fine-grained tactile temporal dynamics during physical interactions. We consider that advancing dynamic tactile perception requires a systematic hierarchy of dynamic perception capabilities to guide both data collection and model design. To address the lack of tactile data with rich dynamic information, we present ToucHD, a large-scale hierarchical tactile dataset spanning tactile atomic actions, real-world manipulations, and touch-force paired data. Beyond scale, ToucHD establishes a comprehensive tactile dynamic data ecosystem that explicitly supports hierarchical perception capabilities from the data perspective. Building on it, we propose AnyTouch 2, a general tactile representation learning framework for diverse optical tactile sensors that unifies object-level understanding with fine-grained, force-aware dynamic perception. The framework captures both pixel-level and action-specific deformations across frames, while explicitly modeling physical force dynamics, thereby learning multi-level dynamic perception capabilities from the model perspective. We evaluate our model on benchmarks that covers static object properties and dynamic physical attributes, as well as real-world manipulation tasks spanning multiple tiers of dynamic perception capabilities-from basic object-level understanding to force-aware dexterous manipulation. Experimental results demonstrate consistent and strong performance across sensors and tasks.
P1-VL: Bridging Visual Perception and Scientific Reasoning in Physics Olympiads
Feb 10, 2026Abstract:The transition from symbolic manipulation to science-grade reasoning represents a pivotal frontier for Large Language Models (LLMs), with physics serving as the critical test anchor for binding abstract logic to physical reality. Physics demands that a model maintain physical consistency with the laws governing the universe, a task that fundamentally requires multimodal perception to ground abstract logic in reality. At the Olympiad level, diagrams are often constitutive rather than illustrative, containing essential constraints, such as boundary conditions and spatial symmetries, that are absent from the text. To bridge this visual-logical gap, we introduce P1-VL, a family of open-source vision-language models engineered for advanced scientific reasoning. Our method harmonizes Curriculum Reinforcement Learning, which employs progressive difficulty expansion to stabilize post-training, with Agentic Augmentation, enabling iterative self-verification at inference. Evaluated on HiPhO, a rigorous benchmark of 13 exams from 2024-2025, our flagship P1-VL-235B-A22B becomes the first open-source Vision-Language Model (VLM) to secure 12 gold medals and achieves the state-of-the-art performance in the open-source models. Our agent-augmented system achieves the No.2 overall rank globally, trailing only Gemini-3-Pro. Beyond physics, P1-VL demonstrates remarkable scientific reasoning capacity and generalizability, establishing significant leads over base models in STEM benchmarks. By open-sourcing P1-VL, we provide a foundational step toward general-purpose physical intelligence to better align visual perceptions with abstract physical laws for machine scientific discovery.
Beyond Static Tools: Test-Time Tool Evolution for Scientific Reasoning
Jan 12, 2026Abstract:The central challenge of AI for Science is not reasoning alone, but the ability to create computational methods in an open-ended scientific world. Existing LLM-based agents rely on static, pre-defined tool libraries, a paradigm that fundamentally fails in scientific domains where tools are sparse, heterogeneous, and intrinsically incomplete. In this paper, we propose Test-Time Tool Evolution (TTE), a new paradigm that enables agents to synthesize, verify, and evolve executable tools during inference. By transforming tools from fixed resources into problem-driven artifacts, TTE overcomes the rigidity and long-tail limitations of static tool libraries. To facilitate rigorous evaluation, we introduce SciEvo, a benchmark comprising 1,590 scientific reasoning tasks supported by 925 automatically evolved tools. Extensive experiments show that TTE achieves state-of-the-art performance in both accuracy and tool efficiency, while enabling effective cross-domain adaptation of computational tools. The code and benchmark have been released at https://github.com/lujiaxuan0520/Test-Time-Tool-Evol.
SketchThinker-R1: Towards Efficient Sketch-Style Reasoning in Large Multimodal Models
Jan 06, 2026Abstract:Despite the empirical success of extensive, step-by-step reasoning in large multimodal models, long reasoning processes inevitably incur substantial computational overhead, i.e., in terms of higher token costs and increased response time, which undermines inference efficiency. In contrast, humans often employ sketch-style reasoning: a concise, goal-directed cognitive process that prioritizes salient information and enables efficient problem-solving. Inspired by this cognitive efficiency, we propose SketchThinker-R1, which incentivizes sketch-style reasoning ability in large multimodal models. Our method consists of three primary stages. In the Sketch-Mode Cold Start stage, we convert standard long reasoning process into sketch-style reasoning and finetune base multimodal model, instilling initial sketch-style reasoning capability. Next, we train SketchJudge Reward Model, which explicitly evaluates thinking process of model and assigns higher scores to sketch-style reasoning. Finally, we conduct Sketch-Thinking Reinforcement Learning under supervision of SketchJudge to further generalize sketch-style reasoning ability. Experimental evaluation on four benchmarks reveals that our SketchThinker-R1 achieves over 64% reduction in reasoning token cost without compromising final answer accuracy. Qualitative analysis further shows that sketch-style reasoning focuses more on key cues during problem solving.
SCP: Accelerating Discovery with a Global Web of Autonomous Scientific Agents
Dec 30, 2025Abstract:We introduce SCP: the Science Context Protocol, an open-source standard designed to accelerate discovery by enabling a global network of autonomous scientific agents. SCP is built on two foundational pillars: (1) Unified Resource Integration: At its core, SCP provides a universal specification for describing and invoking scientific resources, spanning software tools, models, datasets, and physical instruments. This protocol-level standardization enables AI agents and applications to discover, call, and compose capabilities seamlessly across disparate platforms and institutional boundaries. (2) Orchestrated Experiment Lifecycle Management: SCP complements the protocol with a secure service architecture, which comprises a centralized SCP Hub and federated SCP Servers. This architecture manages the complete experiment lifecycle (registration, planning, execution, monitoring, and archival), enforces fine-grained authentication and authorization, and orchestrates traceable, end-to-end workflows that bridge computational and physical laboratories. Based on SCP, we have constructed a scientific discovery platform that offers researchers and agents a large-scale ecosystem of more than 1,600 tool resources. Across diverse use cases, SCP facilitates secure, large-scale collaboration between heterogeneous AI systems and human researchers while significantly reducing integration overhead and enhancing reproducibility. By standardizing scientific context and tool orchestration at the protocol level, SCP establishes essential infrastructure for scalable, multi-institution, agent-driven science.
SciEvalKit: An Open-source Evaluation Toolkit for Scientific General Intelligence
Dec 30, 2025Abstract:We introduce SciEvalKit, a unified benchmarking toolkit designed to evaluate AI models for science across a broad range of scientific disciplines and task capabilities. Unlike general-purpose evaluation platforms, SciEvalKit focuses on the core competencies of scientific intelligence, including Scientific Multimodal Perception, Scientific Multimodal Reasoning, Scientific Multimodal Understanding, Scientific Symbolic Reasoning, Scientific Code Generation, Science Hypothesis Generation and Scientific Knowledge Understanding. It supports six major scientific domains, spanning from physics and chemistry to astronomy and materials science. SciEvalKit builds a foundation of expert-grade scientific benchmarks, curated from real-world, domain-specific datasets, ensuring that tasks reflect authentic scientific challenges. The toolkit features a flexible, extensible evaluation pipeline that enables batch evaluation across models and datasets, supports custom model and dataset integration, and provides transparent, reproducible, and comparable results. By bridging capability-based evaluation and disciplinary diversity, SciEvalKit offers a standardized yet customizable infrastructure to benchmark the next generation of scientific foundation models and intelligent agents. The toolkit is open-sourced and actively maintained to foster community-driven development and progress in AI4Science.
MiST: Understanding the Role of Mid-Stage Scientific Training in Developing Chemical Reasoning Models
Dec 24, 2025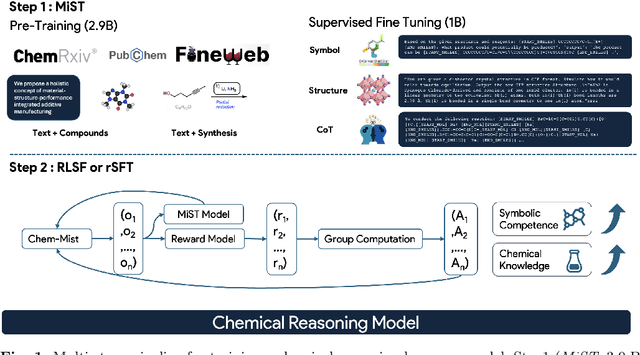
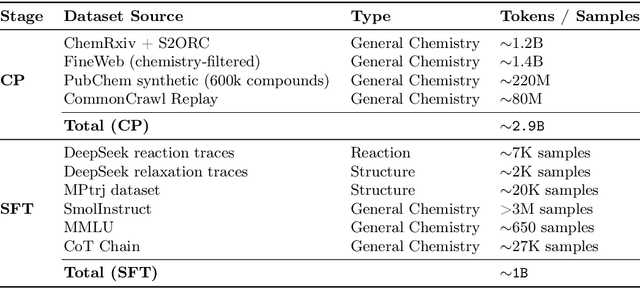
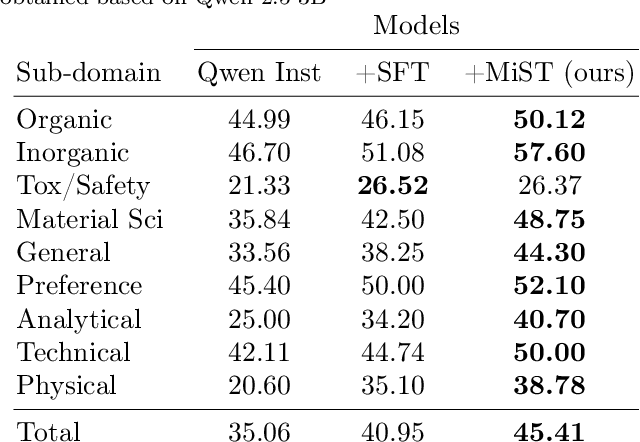
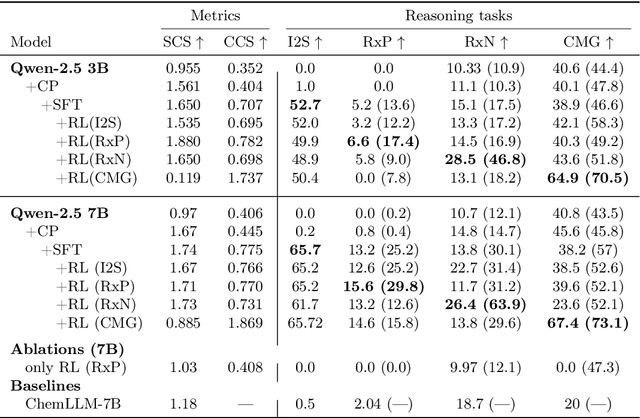
Abstract:Large Language Models can develop reasoning capabilities through online fine-tuning with rule-based rewards. However, recent studies reveal a critical constraint: reinforcement learning succeeds only when the base model already assigns non-negligible probability to correct answers -- a property we term 'latent solvability'. This work investigates the emergence of chemical reasoning capabilities and what these prerequisites mean for chemistry. We identify two necessary conditions for RL-based chemical reasoning: 1) Symbolic competence, and 2) Latent chemical knowledge. We propose mid-stage scientific training (MiST): a set of mid-stage training techniques to satisfy these, including data-mixing with SMILES/CIF-aware pre-processing, continued pre-training on 2.9B tokens, and supervised fine-tuning on 1B tokens. These steps raise the latent-solvability score on 3B and 7B models by up to 1.8x, and enable RL to lift top-1 accuracy from 10.9 to 63.9% on organic reaction naming, and from 40.6 to 67.4% on inorganic material generation. Similar results are observed for other challenging chemical tasks, while producing interpretable reasoning traces. Our results define clear prerequisites for chemical reasoning training and highlight the broader role of mid-stage training in unlocking reasoning capabilities.
An Agentic Framework for Autonomous Materials Computation
Dec 22, 2025



Abstract:Large Language Models (LLMs) have emerged as powerful tools for accelerating scientific discovery, yet their static knowledge and hallucination issues hinder autonomous research applications. Recent advances integrate LLMs into agentic frameworks, enabling retrieval, reasoning, and tool use for complex scientific workflows. Here, we present a domain-specialized agent designed for reliable automation of first-principles materials computations. By embedding domain expertise, the agent ensures physically coherent multi-step workflows and consistently selects convergent, well-posed parameters, thereby enabling reliable end-to-end computational execution. A new benchmark of diverse computational tasks demonstrates that our system significantly outperforms standalone LLMs in both accuracy and robustness. This work establishes a verifiable foundation for autonomous computational experimentation and represents a key step toward fully automated scientific discovery.
Probing Scientific General Intelligence of LLMs with Scientist-Aligned Workflows
Dec 18, 2025Abstract:Despite advances in scientific AI, a coherent framework for Scientific General Intelligence (SGI)-the ability to autonomously conceive, investigate, and reason across scientific domains-remains lacking. We present an operational SGI definition grounded in the Practical Inquiry Model (PIM: Deliberation, Conception, Action, Perception) and operationalize it via four scientist-aligned tasks: deep research, idea generation, dry/wet experiments, and experimental reasoning. SGI-Bench comprises over 1,000 expert-curated, cross-disciplinary samples inspired by Science's 125 Big Questions, enabling systematic evaluation of state-of-the-art LLMs. Results reveal gaps: low exact match (10--20%) in deep research despite step-level alignment; ideas lacking feasibility and detail; high code executability but low execution result accuracy in dry experiments; low sequence fidelity in wet protocols; and persistent multimodal comparative-reasoning challenges. We further introduce Test-Time Reinforcement Learning (TTRL), which optimizes retrieval-augmented novelty rewards at inference, enhancing hypothesis novelty without reference answer. Together, our PIM-grounded definition, workflow-centric benchmark, and empirical insights establish a foundation for AI systems that genuinely participate in scientific discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge