Dingning Liu
3DAxisPrompt: Promoting the 3D Grounding and Reasoning in GPT-4o
Mar 17, 2025Abstract:Multimodal Large Language Models (MLLMs) exhibit impressive capabilities across a variety of tasks, especially when equipped with carefully designed visual prompts. However, existing studies primarily focus on logical reasoning and visual understanding, while the capability of MLLMs to operate effectively in 3D vision remains an ongoing area of exploration. In this paper, we introduce a novel visual prompting method, called 3DAxisPrompt, to elicit the 3D understanding capabilities of MLLMs in real-world scenes. More specifically, our method leverages the 3D coordinate axis and masks generated from the Segment Anything Model (SAM) to provide explicit geometric priors to MLLMs and then extend their impressive 2D grounding and reasoning ability to real-world 3D scenarios. Besides, we first provide a thorough investigation of the potential visual prompting formats and conclude our findings to reveal the potential and limits of 3D understanding capabilities in GPT-4o, as a representative of MLLMs. Finally, we build evaluation environments with four datasets, i.e., ScanRefer, ScanNet, FMB, and nuScene datasets, covering various 3D tasks. Based on this, we conduct extensive quantitative and qualitative experiments, which demonstrate the effectiveness of the proposed method. Overall, our study reveals that MLLMs, with the help of 3DAxisPrompt, can effectively perceive an object's 3D position in real-world scenarios. Nevertheless, a single prompt engineering approach does not consistently achieve the best outcomes for all 3D tasks. This study highlights the feasibility of leveraging MLLMs for 3D vision grounding/reasoning with prompt engineering techniques.
Towards Efficient and Intelligent Laser Weeding: Method and Dataset for Weed Stem Detection
Feb 10, 2025



Abstract:Weed control is a critical challenge in modern agriculture, as weeds compete with crops for essential nutrient resources, significantly reducing crop yield and quality. Traditional weed control methods, including chemical and mechanical approaches, have real-life limitations such as associated environmental impact and efficiency. An emerging yet effective approach is laser weeding, which uses a laser beam as the stem cutter. Although there have been studies that use deep learning in weed recognition, its application in intelligent laser weeding still requires a comprehensive understanding. Thus, this study represents the first empirical investigation of weed recognition for laser weeding. To increase the efficiency of laser beam cut and avoid damaging the crops of interest, the laser beam shall be directly aimed at the weed root. Yet, weed stem detection remains an under-explored problem. We integrate the detection of crop and weed with the localization of weed stem into one end-to-end system. To train and validate the proposed system in a real-life scenario, we curate and construct a high-quality weed stem detection dataset with human annotations. The dataset consists of 7,161 high-resolution pictures collected in the field with annotations of 11,151 instances of weed. Experimental results show that the proposed system improves weeding accuracy by 6.7% and reduces energy cost by 32.3% compared to existing weed recognition systems.
3DAxiesPrompts: Unleashing the 3D Spatial Task Capabilities of GPT-4V
Dec 15, 2023Abstract:In this work, we present a new visual prompting method called 3DAxiesPrompts (3DAP) to unleash the capabilities of GPT-4V in performing 3D spatial tasks. Our investigation reveals that while GPT-4V exhibits proficiency in discerning the position and interrelations of 2D entities through current visual prompting techniques, its abilities in handling 3D spatial tasks have yet to be explored. In our approach, we create a 3D coordinate system tailored to 3D imagery, complete with annotated scale information. By presenting images infused with the 3DAP visual prompt as inputs, we empower GPT-4V to ascertain the spatial positioning information of the given 3D target image with a high degree of precision. Through experiments, We identified three tasks that could be stably completed using the 3DAP method, namely, 2D to 3D Point Reconstruction, 2D to 3D point matching, and 3D Object Detection. We perform experiments on our proposed dataset 3DAP-Data, the results from these experiments validate the efficacy of 3DAP-enhanced GPT-4V inputs, marking a significant stride in 3D spatial task execution.
LAMM: Language-Assisted Multi-Modal Instruction-Tuning Dataset, Framework, and Benchmark
Jun 18, 2023
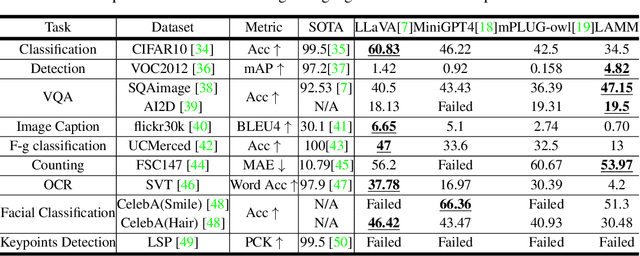
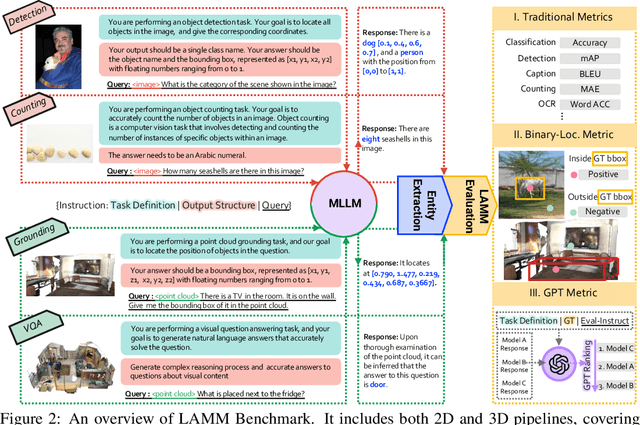
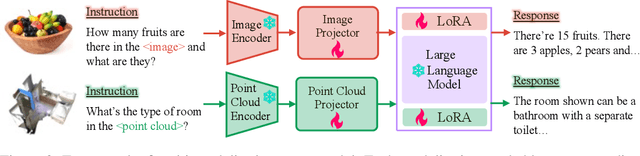
Abstract:Large language models have become a potential pathway toward achieving artificial general intelligence. Recent works on multi-modal large language models have demonstrated their effectiveness in handling visual modalities. In this work, we extend the research of MLLMs to point clouds and present the LAMM-Dataset and LAMM-Benchmark for 2D image and 3D point cloud understanding. We also establish an extensible framework to facilitate the extension of MLLMs to additional modalities. Our main contribution is three-fold: 1) We present the LAMM-Dataset and LAMM-Benchmark, which cover almost all high-level vision tasks for 2D and 3D vision. Extensive experiments validate the effectiveness of our dataset and benchmark. 2) We demonstrate the detailed methods of constructing instruction-tuning datasets and benchmarks for MLLMs, which will enable future research on MLLMs to scale up and extend to other domains, tasks, and modalities faster. 3) We provide a primary but potential MLLM training framework optimized for modalities' extension. We also provide baseline models, comprehensive experimental observations, and analysis to accelerate future research. Codes and datasets are now available at https://github.com/OpenLAMM/LAMM.
Stochastic functional analysis with applications to robust machine learning
Oct 04, 2021
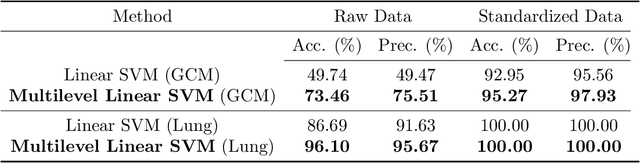
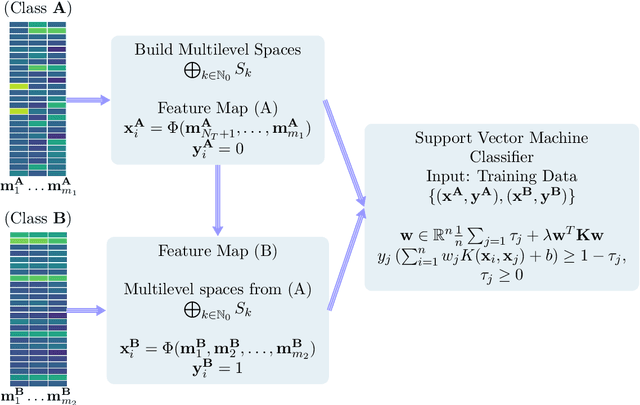

Abstract:It is well-known that machine learning protocols typically under-utilize information on the probability distributions of feature vectors and related data, and instead directly compute regression or classification functions of feature vectors. In this paper we introduce a set of novel features for identifying underlying stochastic behavior of input data using the Karhunen-Lo\'{e}ve (KL) expansion, where classification is treated as detection of anomalies from a (nominal) signal class. These features are constructed from the recent Functional Data Analysis (FDA) theory for anomaly detection. The related signal decomposition is an exact hierarchical tensor product expansion with known optimality properties for approximating stochastic processes (random fields) with finite dimensional function spaces. In principle these primary low dimensional spaces can capture most of the stochastic behavior of `underlying signals' in a given nominal class, and can reject signals in alternative classes as stochastic anomalies. Using a hierarchical finite dimensional KL expansion of the nominal class, a series of orthogonal nested subspaces is constructed for detecting anomalous signal components. Projection coefficients of input data in these subspaces are then used to train an ML classifier. However, due to the split of the signal into nominal and anomalous projection components, clearer separation surfaces of the classes arise. In fact we show that with a sufficiently accurate estimation of the covariance structure of the nominal class, a sharp classification can be obtained. We carefully formulate this concept and demonstrate it on a number of high-dimensional datasets in cancer diagnostics. This method leads to a significant increase in precision and accuracy over the current top benchmarks for the Global Cancer Map (GCM) gene expression network dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge