Greg Corrado
General Geospatial Inference with a Population Dynamics Foundation Model
Nov 13, 2024



Abstract:Supporting the health and well-being of dynamic populations around the world requires governmental agencies, organizations and researchers to understand and reason over complex relationships between human behavior and local contexts in order to identify high-risk groups and strategically allocate limited resources. Traditional approaches to these classes of problems often entail developing manually curated, task-specific features and models to represent human behavior and the natural and built environment, which can be challenging to adapt to new, or even, related tasks. To address this, we introduce a Population Dynamics Foundation Model (PDFM) that aims to capture the relationships between diverse data modalities and is applicable to a broad range of geospatial tasks. We first construct a geo-indexed dataset for postal codes and counties across the United States, capturing rich aggregated information on human behavior from maps, busyness, and aggregated search trends, and environmental factors such as weather and air quality. We then model this data and the complex relationships between locations using a graph neural network, producing embeddings that can be adapted to a wide range of downstream tasks using relatively simple models. We evaluate the effectiveness of our approach by benchmarking it on 27 downstream tasks spanning three distinct domains: health indicators, socioeconomic factors, and environmental measurements. The approach achieves state-of-the-art performance on all 27 geospatial interpolation tasks, and on 25 out of the 27 extrapolation and super-resolution tasks. We combined the PDFM with a state-of-the-art forecasting foundation model, TimesFM, to predict unemployment and poverty, achieving performance that surpasses fully supervised forecasting. The full set of embeddings and sample code are publicly available for researchers.
Advancing Multimodal Medical Capabilities of Gemini
May 06, 2024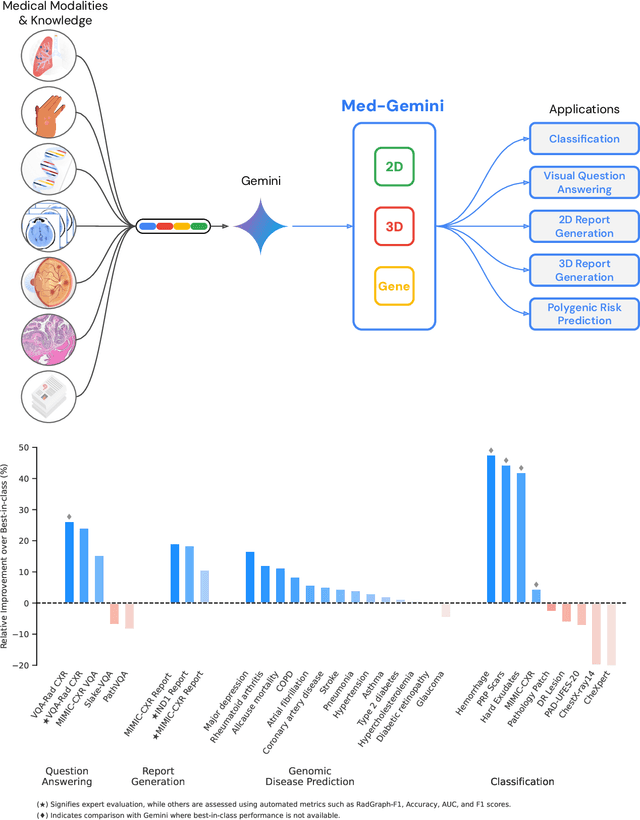
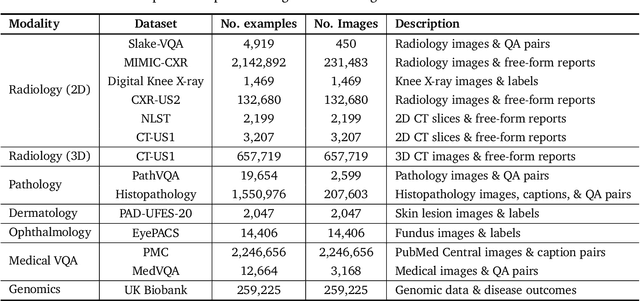

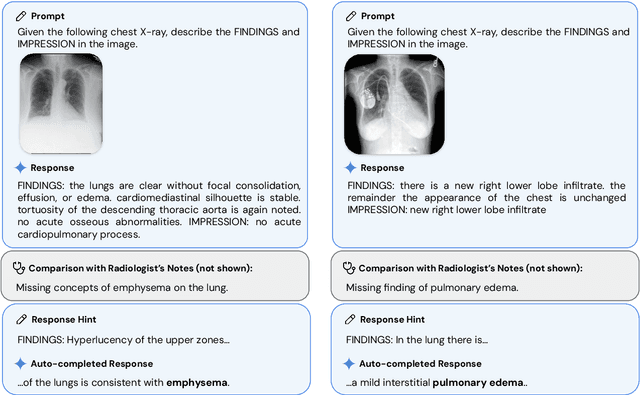
Abstract:Many clinical tasks require an understanding of specialized data, such as medical images and genomics, which is not typically found in general-purpose large multimodal models. Building upon Gemini's multimodal models, we develop several models within the new Med-Gemini family that inherit core capabilities of Gemini and are optimized for medical use via fine-tuning with 2D and 3D radiology, histopathology, ophthalmology, dermatology and genomic data. Med-Gemini-2D sets a new standard for AI-based chest X-ray (CXR) report generation based on expert evaluation, exceeding previous best results across two separate datasets by an absolute margin of 1% and 12%, where 57% and 96% of AI reports on normal cases, and 43% and 65% on abnormal cases, are evaluated as "equivalent or better" than the original radiologists' reports. We demonstrate the first ever large multimodal model-based report generation for 3D computed tomography (CT) volumes using Med-Gemini-3D, with 53% of AI reports considered clinically acceptable, although additional research is needed to meet expert radiologist reporting quality. Beyond report generation, Med-Gemini-2D surpasses the previous best performance in CXR visual question answering (VQA) and performs well in CXR classification and radiology VQA, exceeding SoTA or baselines on 17 of 20 tasks. In histopathology, ophthalmology, and dermatology image classification, Med-Gemini-2D surpasses baselines across 18 out of 20 tasks and approaches task-specific model performance. Beyond imaging, Med-Gemini-Polygenic outperforms the standard linear polygenic risk score-based approach for disease risk prediction and generalizes to genetically correlated diseases for which it has never been trained. Although further development and evaluation are necessary in the safety-critical medical domain, our results highlight the potential of Med-Gemini across a wide range of medical tasks.
Capabilities of Gemini Models in Medicine
May 01, 2024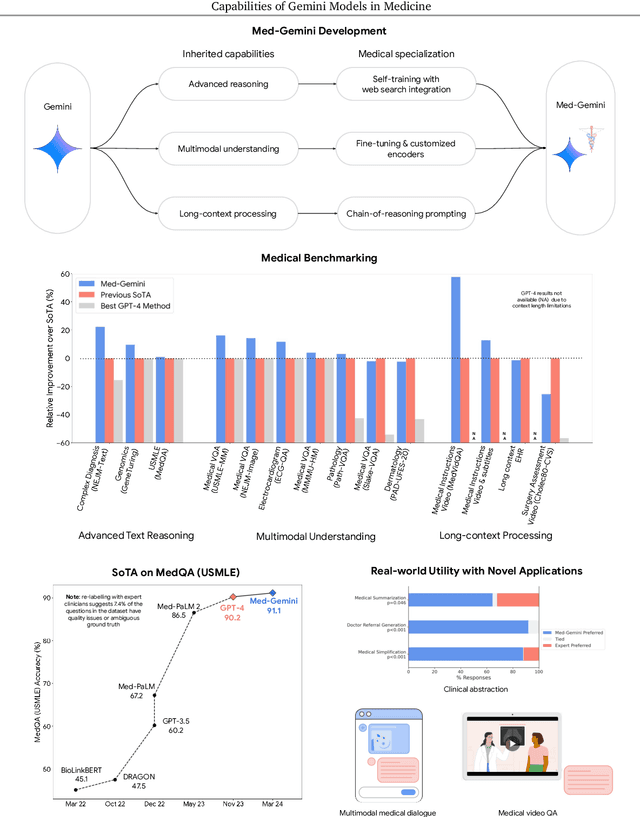
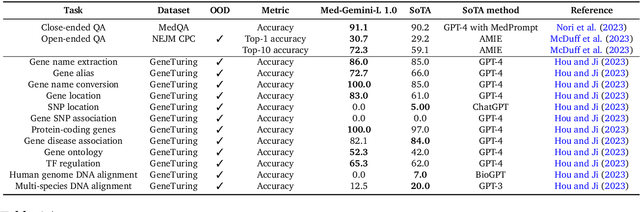
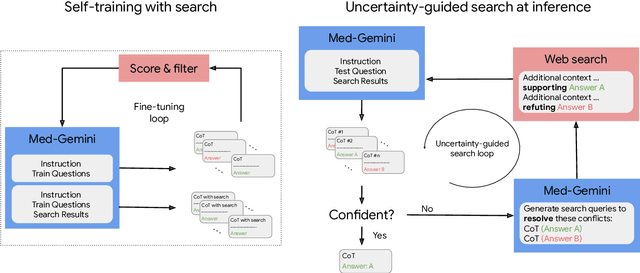
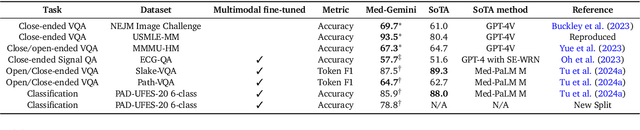
Abstract:Excellence in a wide variety of medical applications poses considerable challenges for AI, requiring advanced reasoning, access to up-to-date medical knowledge and understanding of complex multimodal data. Gemini models, with strong general capabilities in multimodal and long-context reasoning, offer exciting possibilities in medicine. Building on these core strengths of Gemini, we introduce Med-Gemini, a family of highly capable multimodal models that are specialized in medicine with the ability to seamlessly use web search, and that can be efficiently tailored to novel modalities using custom encoders. We evaluate Med-Gemini on 14 medical benchmarks, establishing new state-of-the-art (SoTA) performance on 10 of them, and surpass the GPT-4 model family on every benchmark where a direct comparison is viable, often by a wide margin. On the popular MedQA (USMLE) benchmark, our best-performing Med-Gemini model achieves SoTA performance of 91.1% accuracy, using a novel uncertainty-guided search strategy. On 7 multimodal benchmarks including NEJM Image Challenges and MMMU (health & medicine), Med-Gemini improves over GPT-4V by an average relative margin of 44.5%. We demonstrate the effectiveness of Med-Gemini's long-context capabilities through SoTA performance on a needle-in-a-haystack retrieval task from long de-identified health records and medical video question answering, surpassing prior bespoke methods using only in-context learning. Finally, Med-Gemini's performance suggests real-world utility by surpassing human experts on tasks such as medical text summarization, alongside demonstrations of promising potential for multimodal medical dialogue, medical research and education. Taken together, our results offer compelling evidence for Med-Gemini's potential, although further rigorous evaluation will be crucial before real-world deployment in this safety-critical domain.
A Toolbox for Surfacing Health Equity Harms and Biases in Large Language Models
Mar 18, 2024Abstract:Large language models (LLMs) hold immense promise to serve complex health information needs but also have the potential to introduce harm and exacerbate health disparities. Reliably evaluating equity-related model failures is a critical step toward developing systems that promote health equity. In this work, we present resources and methodologies for surfacing biases with potential to precipitate equity-related harms in long-form, LLM-generated answers to medical questions and then conduct an empirical case study with Med-PaLM 2, resulting in the largest human evaluation study in this area to date. Our contributions include a multifactorial framework for human assessment of LLM-generated answers for biases, and EquityMedQA, a collection of seven newly-released datasets comprising both manually-curated and LLM-generated questions enriched for adversarial queries. Both our human assessment framework and dataset design process are grounded in an iterative participatory approach and review of possible biases in Med-PaLM 2 answers to adversarial queries. Through our empirical study, we find that the use of a collection of datasets curated through a variety of methodologies, coupled with a thorough evaluation protocol that leverages multiple assessment rubric designs and diverse rater groups, surfaces biases that may be missed via narrower evaluation approaches. Our experience underscores the importance of using diverse assessment methodologies and involving raters of varying backgrounds and expertise. We emphasize that while our framework can identify specific forms of bias, it is not sufficient to holistically assess whether the deployment of an AI system promotes equitable health outcomes. We hope the broader community leverages and builds on these tools and methods towards realizing a shared goal of LLMs that promote accessible and equitable healthcare for all.
Multimodal LLMs for health grounded in individual-specific data
Jul 20, 2023



Abstract:Foundation large language models (LLMs) have shown an impressive ability to solve tasks across a wide range of fields including health. To effectively solve personalized health tasks, LLMs need the ability to ingest a diversity of data modalities that are relevant to an individual's health status. In this paper, we take a step towards creating multimodal LLMs for health that are grounded in individual-specific data by developing a framework (HeLM: Health Large Language Model for Multimodal Understanding) that enables LLMs to use high-dimensional clinical modalities to estimate underlying disease risk. HeLM encodes complex data modalities by learning an encoder that maps them into the LLM's token embedding space and for simple modalities like tabular data by serializing the data into text. Using data from the UK Biobank, we show that HeLM can effectively use demographic and clinical features in addition to high-dimensional time-series data to estimate disease risk. For example, HeLM achieves an AUROC of 0.75 for asthma prediction when combining tabular and spirogram data modalities compared with 0.49 when only using tabular data. Overall, we find that HeLM outperforms or performs at parity with classical machine learning approaches across a selection of eight binary traits. Furthermore, we investigate the downstream uses of this model such as its generalizability to out-of-distribution traits and its ability to power conversations around individual health and wellness.
AI system for fetal ultrasound in low-resource settings
Mar 18, 2022



Abstract:Despite considerable progress in maternal healthcare, maternal and perinatal deaths remain high in low-to-middle income countries. Fetal ultrasound is an important component of antenatal care, but shortage of adequately trained healthcare workers has limited its adoption. We developed and validated an artificial intelligence (AI) system that uses novice-acquired "blind sweep" ultrasound videos to estimate gestational age (GA) and fetal malpresentation. We further addressed obstacles that may be encountered in low-resourced settings. Using a simplified sweep protocol with real-time AI feedback on sweep quality, we have demonstrated the generalization of model performance to minimally trained novice ultrasound operators using low cost ultrasound devices with on-device AI integration. The GA model was non-inferior to standard fetal biometry estimates with as few as two sweeps, and the fetal malpresentation model had high AUC-ROCs across operators and devices. Our AI models have the potential to assist in upleveling the capabilities of lightly trained ultrasound operators in low resource settings.
Scientific Discovery by Generating Counterfactuals using Image Translation
Jul 10, 2020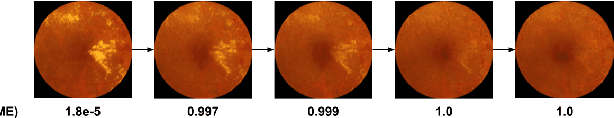
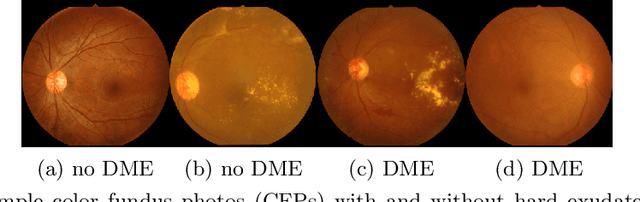
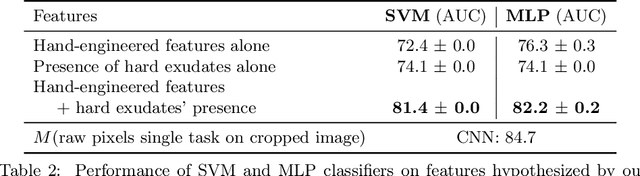
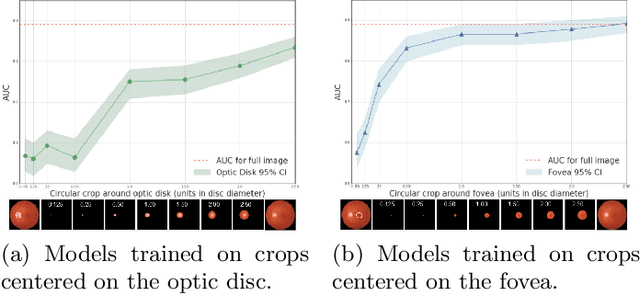
Abstract:Model explanation techniques play a critical role in understanding the source of a model's performance and making its decisions transparent. Here we investigate if explanation techniques can also be used as a mechanism for scientific discovery. We make three contributions: first, we propose a framework to convert predictions from explanation techniques to a mechanism of discovery. Second, we show how generative models in combination with black-box predictors can be used to generate hypotheses (without human priors) that can be critically examined. Third, with these techniques we study classification models for retinal images predicting Diabetic Macular Edema (DME), where recent work showed that a CNN trained on these images is likely learning novel features in the image. We demonstrate that the proposed framework is able to explain the underlying scientific mechanism, thus bridging the gap between the model's performance and human understanding.
* Accepted at MICCAI 2020. This version combines camera-ready and supplement
Detecting Deficient Coverage in Colonoscopies
Jan 26, 2020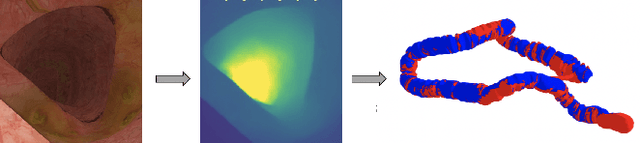
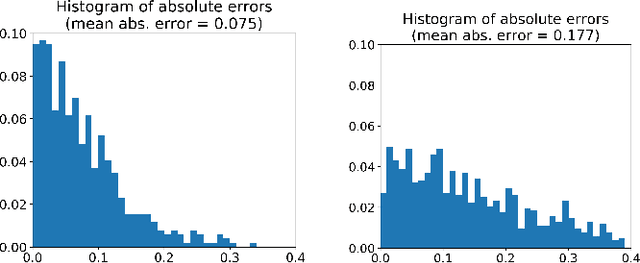
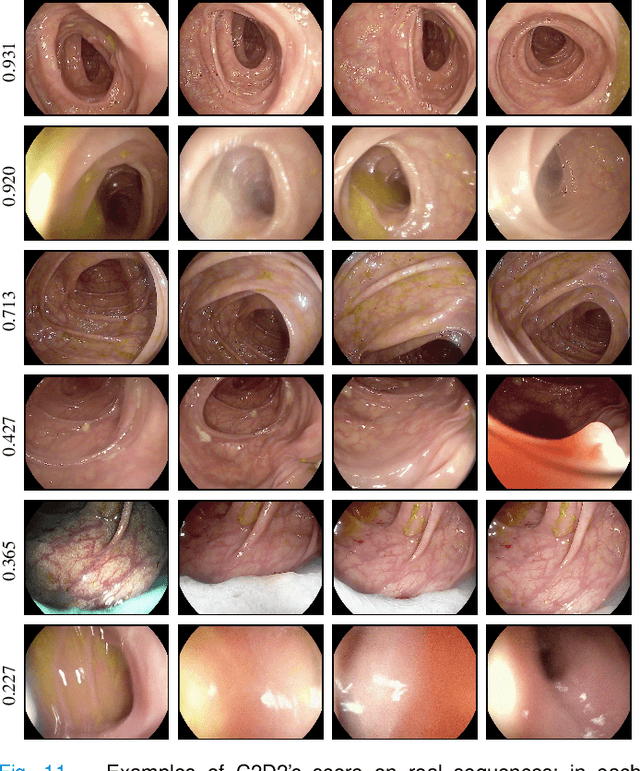
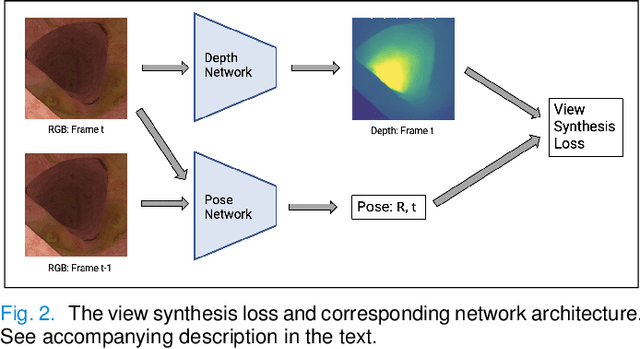
Abstract:Colorectal Cancer (CRC) is a global health problem, resulting in 900K deaths per year. Colonoscopy is the tool of choice for preventing CRC, by detecting polyps before they become cancerous, and removing them. However, colonoscopy is hampered by the fact that endoscopists routinely miss an average of 22-28% of polyps. While some of these missed polyps appear in the endoscopist's field of view, others are missed simply because of substandard coverage of the procedure, i.e. not all of the colon is seen. This paper attempts to rectify the problem of substandard coverage in colonoscopy through the introduction of the C2D2 (Colonoscopy Coverage Deficiency via Depth) algorithm which detects deficient coverage, and can thereby alert the endoscopist to revisit a given area. More specifically, C2D2 consists of two separate algorithms: the first performs depth estimation of the colon given an ordinary RGB video stream; while the second computes coverage given these depth estimates. Rather than compute coverage for the entire colon, our algorithm computes coverage locally, on a segment-by-segment basis; C2D2 can then indicate in real-time whether a particular area of the colon has suffered from deficient coverage, and if so the endoscopist can return to that area. Our coverage algorithm is the first such algorithm to be evaluated in a large-scale way; while our depth estimation technique is the first calibration-free unsupervised method applied to colonoscopies. The C2D2 algorithm achieves state of the art results in the detection of deficient coverage: it is 2.4 times more accurate than human experts.
Explaining an increase in predicted risk for clinical alerts
Jul 10, 2019
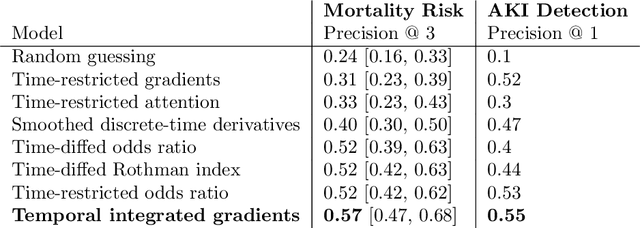

Abstract:Much work aims to explain a model's prediction on a static input. We consider explanations in a temporal setting where a stateful dynamical model produces a sequence of risk estimates given an input at each time step. When the estimated risk increases, the goal of the explanation is to attribute the increase to a few relevant inputs from the past. While our formal setup and techniques are general, we carry out an in-depth case study in a clinical setting. The goal here is to alert a clinician when a patient's risk of deterioration rises. The clinician then has to decide whether to intervene and adjust the treatment. Given a potentially long sequence of new events since she last saw the patient, a concise explanation helps her to quickly triage the alert. We develop methods to lift static attribution techniques to the dynamical setting, where we identify and address challenges specific to dynamics. We then experimentally assess the utility of different explanations of clinical alerts through expert evaluation.
The Algorithmic Automation Problem: Prediction, Triage, and Human Effort
Mar 28, 2019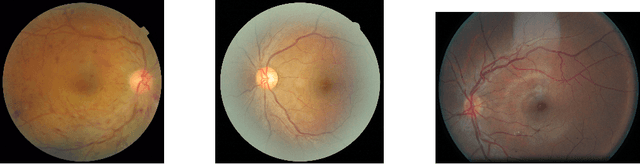
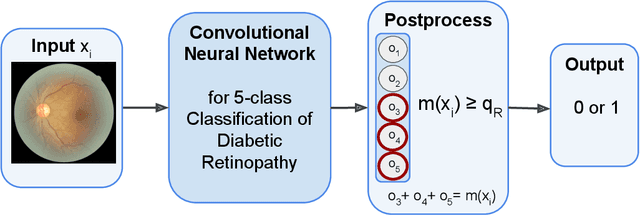
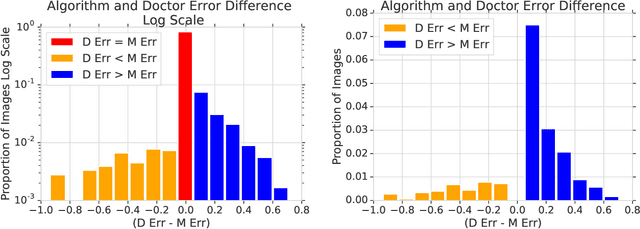
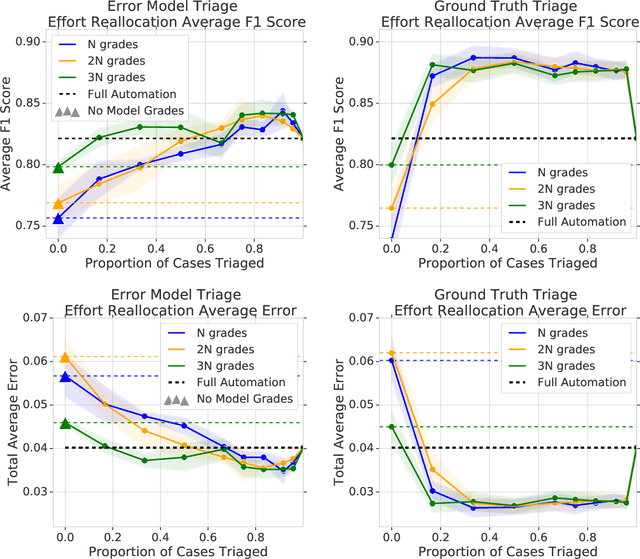
Abstract:In a wide array of areas, algorithms are matching and surpassing the performance of human experts, leading to consideration of the roles of human judgment and algorithmic prediction in these domains. The discussion around these developments, however, has implicitly equated the specific task of prediction with the general task of automation. We argue here that automation is broader than just a comparison of human versus algorithmic performance on a task; it also involves the decision of which instances of the task to give to the algorithm in the first place. We develop a general framework that poses this latter decision as an optimization problem, and we show how basic heuristics for this optimization problem can lead to performance gains even on heavily-studied applications of AI in medicine. Our framework also serves to highlight how effective automation depends crucially on estimating both algorithmic and human error on an instance-by-instance basis, and our results show how improvements in these error estimation problems can yield significant gains for automation as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge