Pinal Bavishi
Robust and Efficient Medical Imaging with Self-Supervision
May 19, 2022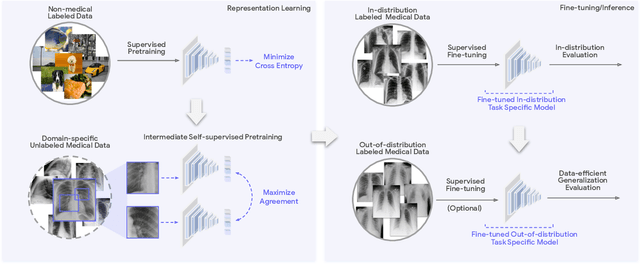
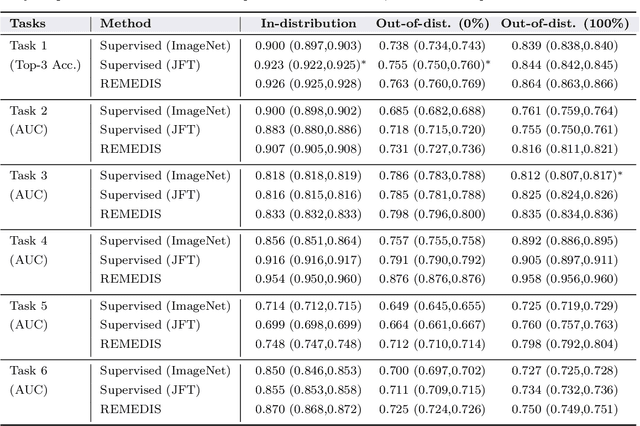
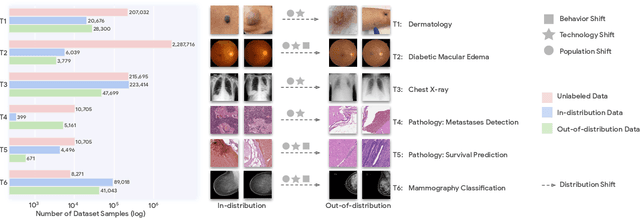
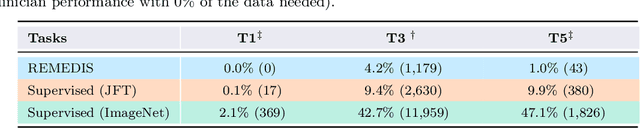
Abstract:Recent progress in Medical Artificial Intelligence (AI) has delivered systems that can reach clinical expert level performance. However, such systems tend to demonstrate sub-optimal "out-of-distribution" performance when evaluated in clinical settings different from the training environment. A common mitigation strategy is to develop separate systems for each clinical setting using site-specific data [1]. However, this quickly becomes impractical as medical data is time-consuming to acquire and expensive to annotate [2]. Thus, the problem of "data-efficient generalization" presents an ongoing difficulty for Medical AI development. Although progress in representation learning shows promise, their benefits have not been rigorously studied, specifically for out-of-distribution settings. To meet these challenges, we present REMEDIS, a unified representation learning strategy to improve robustness and data-efficiency of medical imaging AI. REMEDIS uses a generic combination of large-scale supervised transfer learning with self-supervised learning and requires little task-specific customization. We study a diverse range of medical imaging tasks and simulate three realistic application scenarios using retrospective data. REMEDIS exhibits significantly improved in-distribution performance with up to 11.5% relative improvement in diagnostic accuracy over a strong supervised baseline. More importantly, our strategy leads to strong data-efficient generalization of medical imaging AI, matching strong supervised baselines using between 1% to 33% of retraining data across tasks. These results suggest that REMEDIS can significantly accelerate the life-cycle of medical imaging AI development thereby presenting an important step forward for medical imaging AI to deliver broad impact.
Predicting Risk of Developing Diabetic Retinopathy using Deep Learning
Aug 10, 2020

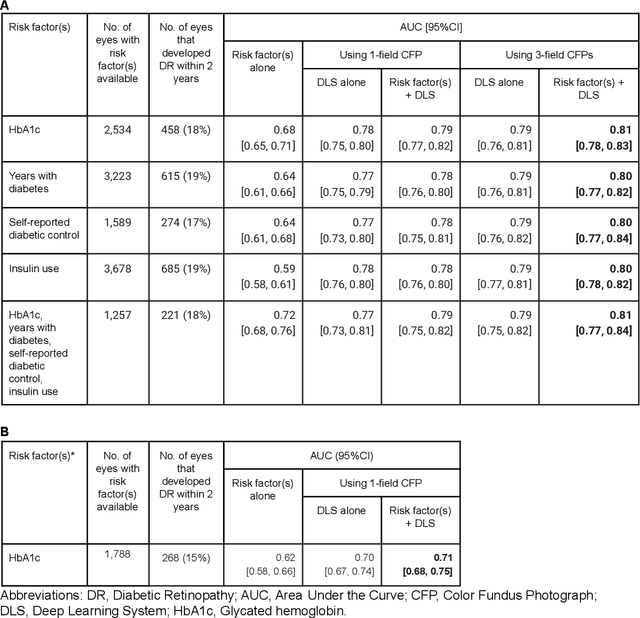

Abstract:Diabetic retinopathy (DR) screening is instrumental in preventing blindness, but faces a scaling challenge as the number of diabetic patients rises. Risk stratification for the development of DR may help optimize screening intervals to reduce costs while improving vision-related outcomes. We created and validated two versions of a deep learning system (DLS) to predict the development of mild-or-worse ("Mild+") DR in diabetic patients undergoing DR screening. The two versions used either three-fields or a single field of color fundus photographs (CFPs) as input. The training set was derived from 575,431 eyes, of which 28,899 had known 2-year outcome, and the remaining were used to augment the training process via multi-task learning. Validation was performed on both an internal validation set (set A; 7,976 eyes; 3,678 with known outcome) and an external validation set (set B; 4,762 eyes; 2,345 with known outcome). For predicting 2-year development of DR, the 3-field DLS had an area under the receiver operating characteristic curve (AUC) of 0.79 (95%CI, 0.78-0.81) on validation set A. On validation set B (which contained only a single field), the 1-field DLS's AUC was 0.70 (95%CI, 0.67-0.74). The DLS was prognostic even after adjusting for available risk factors (p<0.001). When added to the risk factors, the 3-field DLS improved the AUC from 0.72 (95%CI, 0.68-0.76) to 0.81 (95%CI, 0.77-0.84) in validation set A, and the 1-field DLS improved the AUC from 0.62 (95%CI, 0.58-0.66) to 0.71 (95%CI, 0.68-0.75) in validation set B. The DLSs in this study identified prognostic information for DR development from CFPs. This information is independent of and more informative than the available risk factors.
Scientific Discovery by Generating Counterfactuals using Image Translation
Jul 10, 2020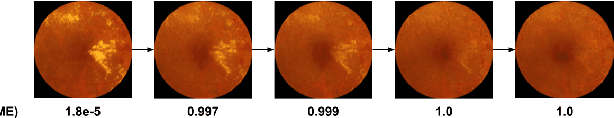
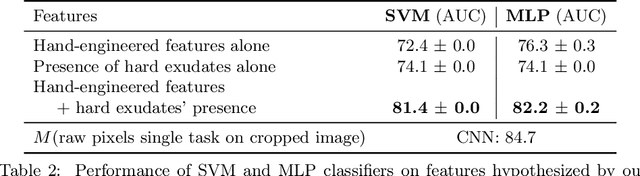
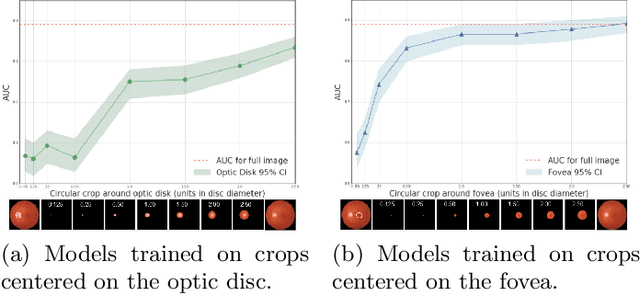
Abstract:Model explanation techniques play a critical role in understanding the source of a model's performance and making its decisions transparent. Here we investigate if explanation techniques can also be used as a mechanism for scientific discovery. We make three contributions: first, we propose a framework to convert predictions from explanation techniques to a mechanism of discovery. Second, we show how generative models in combination with black-box predictors can be used to generate hypotheses (without human priors) that can be critically examined. Third, with these techniques we study classification models for retinal images predicting Diabetic Macular Edema (DME), where recent work showed that a CNN trained on these images is likely learning novel features in the image. We demonstrate that the proposed framework is able to explain the underlying scientific mechanism, thus bridging the gap between the model's performance and human understanding.
* Accepted at MICCAI 2020. This version combines camera-ready and supplement
Predicting optical coherence tomography-derived diabetic macular edema grades from fundus photographs using deep learning
Oct 18, 2018
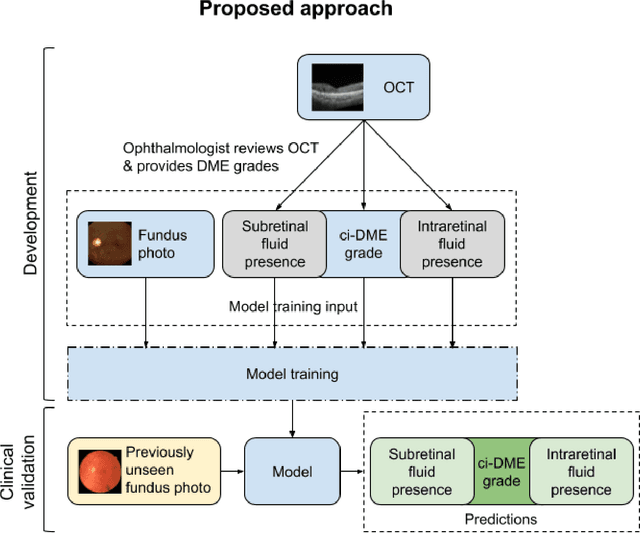
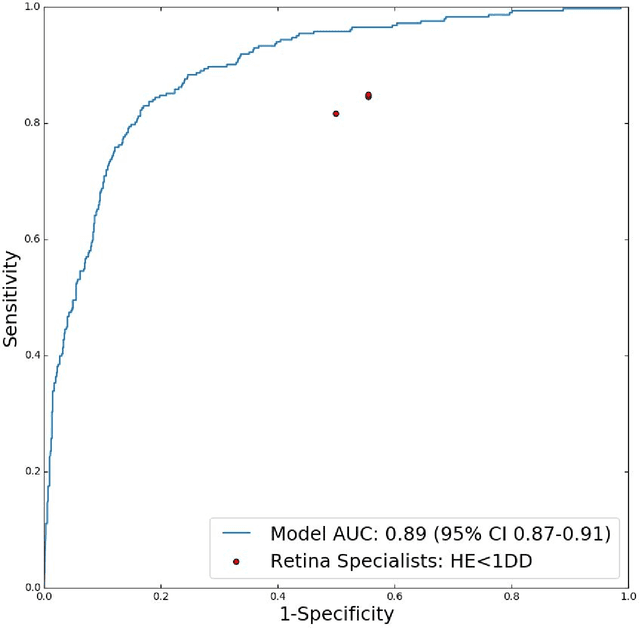
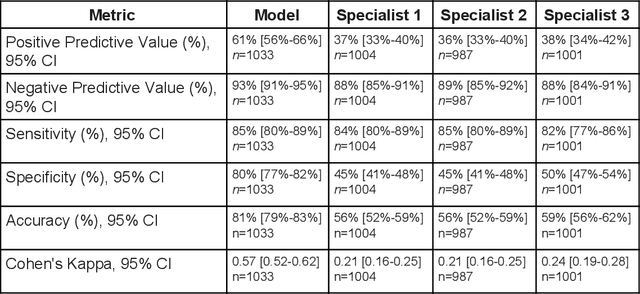
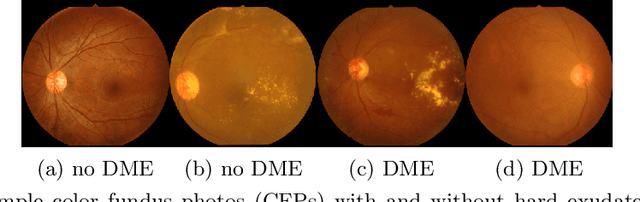
Abstract:Diabetic eye disease is one of the fastest growing causes of preventable blindness. With the advent of anti-VEGF (vascular endothelial growth factor) therapies, it has become increasingly important to detect center-involved diabetic macular edema. However, center-involved diabetic macular edema is diagnosed using optical coherence tomography (OCT), which is not generally available at screening sites because of cost and workflow constraints. Instead, screening programs rely on the detection of hard exudates as a proxy for DME on color fundus photographs, often resulting in high false positive or false negative calls. To improve the accuracy of DME screening, we trained a deep learning model to use color fundus photographs to predict DME grades derived from OCT exams. Our "OCT-DME" model had an AUC of 0.89 (95% CI: 0.87-0.91), which corresponds to a sensitivity of 85% at a specificity of 80%. In comparison, three retinal specialists had similar sensitivities (82-85%), but only half the specificity (45-50%, p<0.001 for each comparison with model). The positive predictive value (PPV) of the OCT-DME model was 61% (95% CI: 56-66%), approximately double the 36-38% by the retina specialists. In addition, we used saliency and other techniques to examine how the model is making its prediction. The ability of deep learning algorithms to make clinically relevant predictions that generally require sophisticated 3D-imaging equipment from simple 2D images has broad relevance to many other applications in medical imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge