Avinash V. Varadarajan
Discovering novel systemic biomarkers in photos of the external eye
Jul 19, 2022
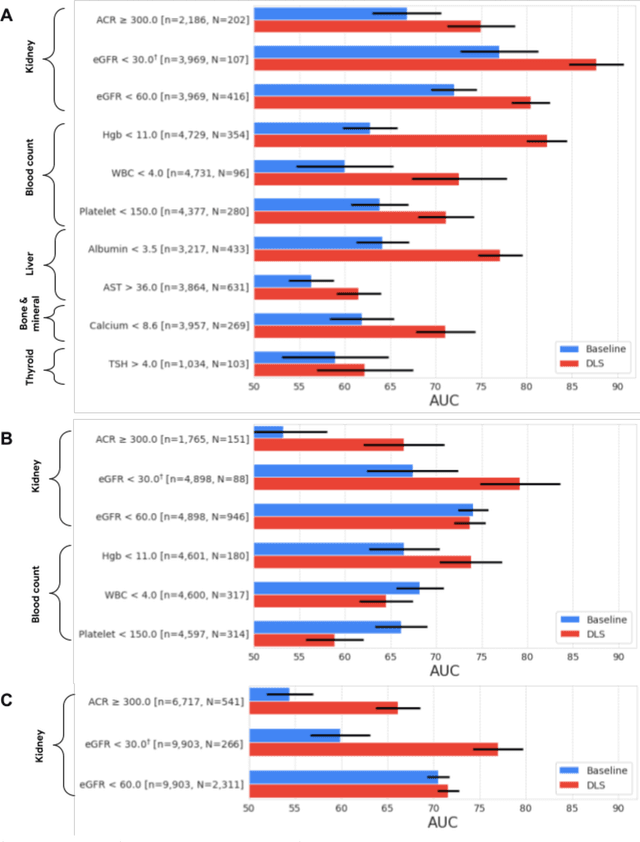
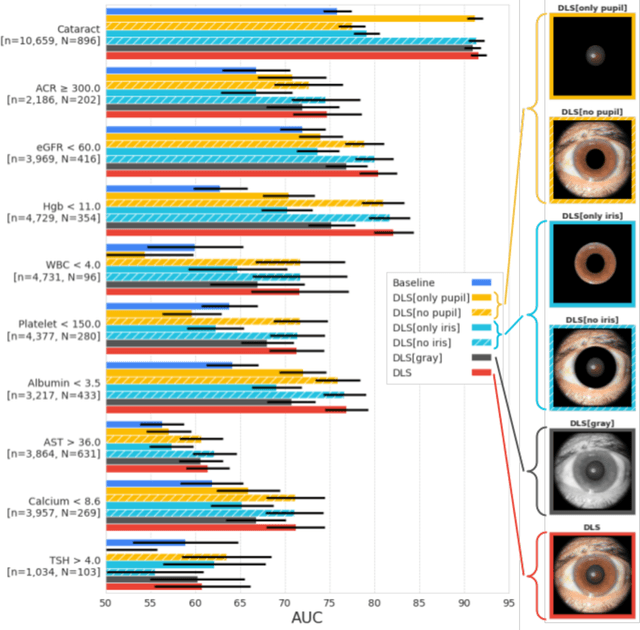
Abstract:External eye photos were recently shown to reveal signs of diabetic retinal disease and elevated HbA1c. In this paper, we evaluate if external eye photos contain information about additional systemic medical conditions. We developed a deep learning system (DLS) that takes external eye photos as input and predicts multiple systemic parameters, such as those related to the liver (albumin, AST); kidney (eGFR estimated using the race-free 2021 CKD-EPI creatinine equation, the urine ACR); bone & mineral (calcium); thyroid (TSH); and blood count (Hgb, WBC, platelets). Development leveraged 151,237 images from 49,015 patients with diabetes undergoing diabetic eye screening in 11 sites across Los Angeles county, CA. Evaluation focused on 9 pre-specified systemic parameters and leveraged 3 validation sets (A, B, C) spanning 28,869 patients with and without diabetes undergoing eye screening in 3 independent sites in Los Angeles County, CA, and the greater Atlanta area, GA. We compared against baseline models incorporating available clinicodemographic variables (e.g. age, sex, race/ethnicity, years with diabetes). Relative to the baseline, the DLS achieved statistically significant superior performance at detecting AST>36, calcium<8.6, eGFR<60, Hgb<11, platelets<150, ACR>=300, and WBC<4 on validation set A (a patient population similar to the development sets), where the AUC of DLS exceeded that of the baseline by 5.2-19.4%. On validation sets B and C, with substantial patient population differences compared to the development sets, the DLS outperformed the baseline for ACR>=300 and Hgb<11 by 7.3-13.2%. Our findings provide further evidence that external eye photos contain important biomarkers of systemic health spanning multiple organ systems. Further work is needed to investigate whether and how these biomarkers can be translated into clinical impact.
Scientific Discovery by Generating Counterfactuals using Image Translation
Jul 10, 2020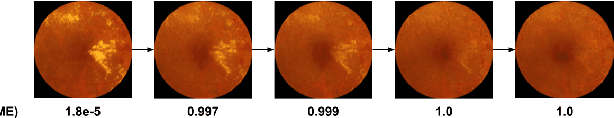
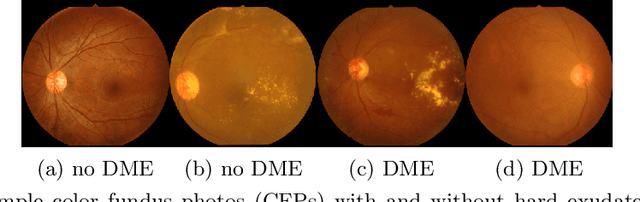
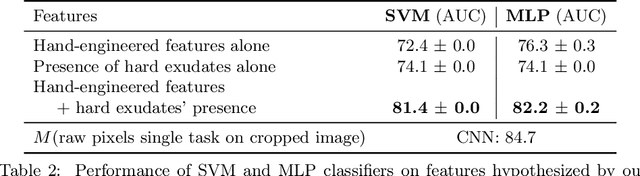
Abstract:Model explanation techniques play a critical role in understanding the source of a model's performance and making its decisions transparent. Here we investigate if explanation techniques can also be used as a mechanism for scientific discovery. We make three contributions: first, we propose a framework to convert predictions from explanation techniques to a mechanism of discovery. Second, we show how generative models in combination with black-box predictors can be used to generate hypotheses (without human priors) that can be critically examined. Third, with these techniques we study classification models for retinal images predicting Diabetic Macular Edema (DME), where recent work showed that a CNN trained on these images is likely learning novel features in the image. We demonstrate that the proposed framework is able to explain the underlying scientific mechanism, thus bridging the gap between the model's performance and human understanding.
* Accepted at MICCAI 2020. This version combines camera-ready and supplement
Detecting Anemia from Retinal Fundus Images
Apr 12, 2019



Abstract:Despite its high prevalence, anemia is often undetected due to the invasiveness and cost of screening and diagnostic tests. Though some non-invasive approaches have been developed, they are less accurate than invasive methods, resulting in an unmet need for more accurate non-invasive methods. Here, we show that deep learning-based algorithms can detect anemia and quantify several related blood measurements using retinal fundus images both in isolation and in combination with basic metadata such as patient demographics. On a validation dataset of 11,388 patients from the UK Biobank, our algorithms achieved a mean absolute error of 0.63 g/dL (95% confidence interval (CI) 0.62-0.64) in quantifying hemoglobin concentration and an area under receiver operating characteristic curve (AUC) of 0.88 (95% CI 0.86-0.89) in detecting anemia. This work shows the potential of automated non-invasive anemia screening based on fundus images, particularly in diabetic patients, who may have regular retinal imaging and are at increased risk of further morbidity and mortality from anemia.
Predicting Progression of Age-related Macular Degeneration from Fundus Images using Deep Learning
Apr 10, 2019



Abstract:Background: Patients with neovascular age-related macular degeneration (AMD) can avoid vision loss via certain therapy. However, methods to predict the progression to neovascular age-related macular degeneration (nvAMD) are lacking. Purpose: To develop and validate a deep learning (DL) algorithm to predict 1-year progression of eyes with no, early, or intermediate AMD to nvAMD, using color fundus photographs (CFP). Design: Development and validation of a DL algorithm. Methods: We trained a DL algorithm to predict 1-year progression to nvAMD, and used 10-fold cross-validation to evaluate this approach on two groups of eyes in the Age-Related Eye Disease Study (AREDS): none/early/intermediate AMD, and intermediate AMD (iAMD) only. We compared the DL algorithm to the manually graded 4-category and 9-step scales in the AREDS dataset. Main outcome measures: Performance of the DL algorithm was evaluated using the sensitivity at 80% specificity for progression to nvAMD. Results: The DL algorithm's sensitivity for predicting progression to nvAMD from none/early/iAMD (78+/-6%) was higher than manual grades from the 9-step scale (67+/-8%) or the 4-category scale (48+/-3%). For predicting progression specifically from iAMD, the DL algorithm's sensitivity (57+/-6%) was also higher compared to the 9-step grades (36+/-8%) and the 4-category grades (20+/-0%). Conclusions: Our DL algorithm performed better in predicting progression to nvAMD than manual grades. Future investigations are required to test the application of this DL algorithm in a real-world clinical setting.
Deep learning for predicting refractive error from retinal fundus images
Dec 21, 2017

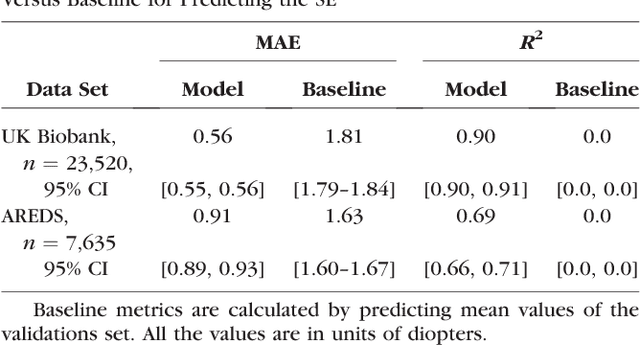
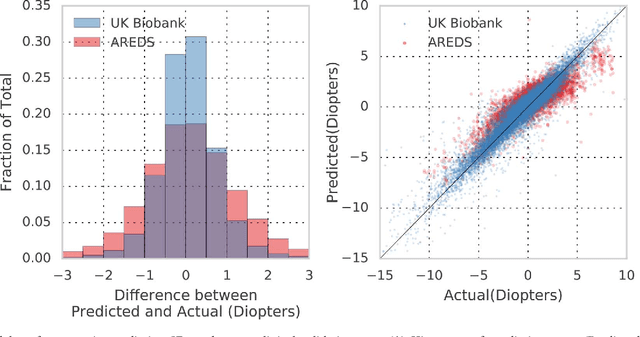
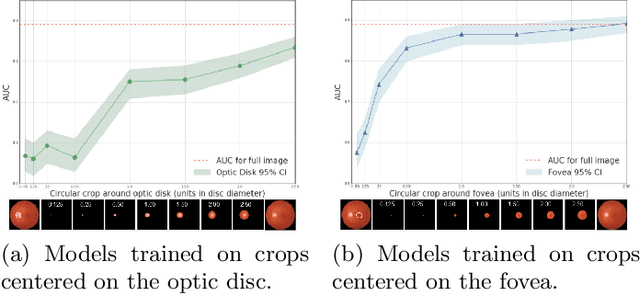
Abstract:Refractive error, one of the leading cause of visual impairment, can be corrected by simple interventions like prescribing eyeglasses. We trained a deep learning algorithm to predict refractive error from the fundus photographs from participants in the UK Biobank cohort, which were 45 degree field of view images and the AREDS clinical trial, which contained 30 degree field of view images. Our model use the "attention" method to identify features that are correlated with refractive error. Mean absolute error (MAE) of the algorithm's prediction compared to the refractive error obtained in the AREDS and UK Biobank. The resulting algorithm had a MAE of 0.56 diopters (95% CI: 0.55-0.56) for estimating spherical equivalent on the UK Biobank dataset and 0.91 diopters (95% CI: 0.89-0.92) for the AREDS dataset. The baseline expected MAE (obtained by simply predicting the mean of this population) was 1.81 diopters (95% CI: 1.79-1.84) for UK Biobank and 1.63 (95% CI: 1.60-1.67) for AREDS. Attention maps suggested that the foveal region was one of the most important areas used by the algorithm to make this prediction, though other regions also contribute to the prediction. The ability to estimate refractive error with high accuracy from retinal fundus photos has not been previously known and demonstrates that deep learning can be applied to make novel predictions from medical images. Given that several groups have recently shown that it is feasible to obtain retinal fundus photos using mobile phones and inexpensive attachments, this work may be particularly relevant in regions of the world where autorefractors may not be readily available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge