Naama Hammel
Using generative AI to investigate medical imagery models and datasets
Jun 01, 2023



Abstract:AI models have shown promise in many medical imaging tasks. However, our ability to explain what signals these models have learned is severely lacking. Explanations are needed in order to increase the trust in AI-based models, and could enable novel scientific discovery by uncovering signals in the data that are not yet known to experts. In this paper, we present a method for automatic visual explanations leveraging team-based expertise by generating hypotheses of what visual signals in the images are correlated with the task. We propose the following 4 steps: (i) Train a classifier to perform a given task (ii) Train a classifier guided StyleGAN-based image generator (StylEx) (iii) Automatically detect and visualize the top visual attributes that the classifier is sensitive towards (iv) Formulate hypotheses for the underlying mechanisms, to stimulate future research. Specifically, we present the discovered attributes to an interdisciplinary panel of experts so that hypotheses can account for social and structural determinants of health. We demonstrate results on eight prediction tasks across three medical imaging modalities: retinal fundus photographs, external eye photographs, and chest radiographs. We showcase examples of attributes that capture clinically known features, confounders that arise from factors beyond physiological mechanisms, and reveal a number of physiologically plausible novel attributes. Our approach has the potential to enable researchers to better understand, improve their assessment, and extract new knowledge from AI-based models. Importantly, we highlight that attributes generated by our framework can capture phenomena beyond physiology or pathophysiology, reflecting the real world nature of healthcare delivery and socio-cultural factors. Finally, we intend to release code to enable researchers to train their own StylEx models and analyze their predictive tasks.
Discovering novel systemic biomarkers in photos of the external eye
Jul 19, 2022
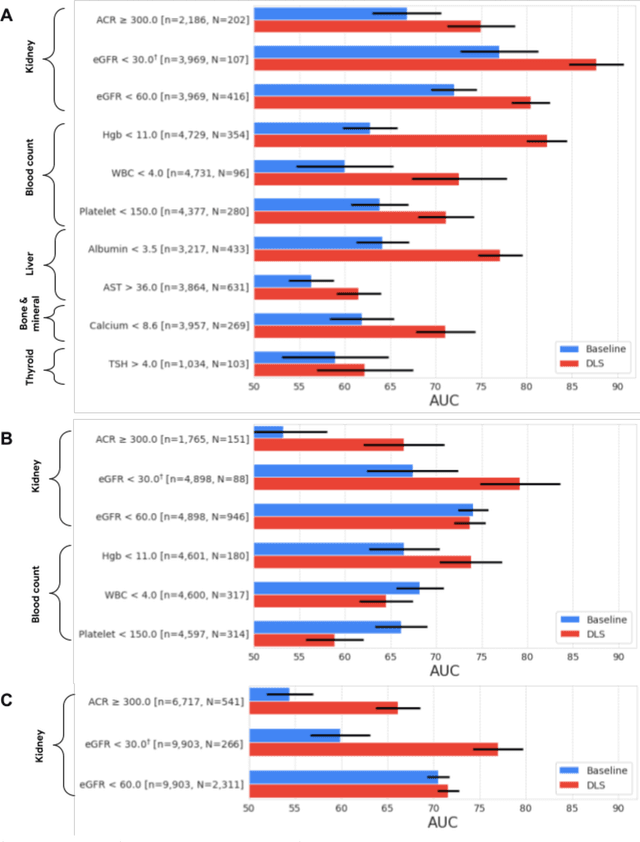
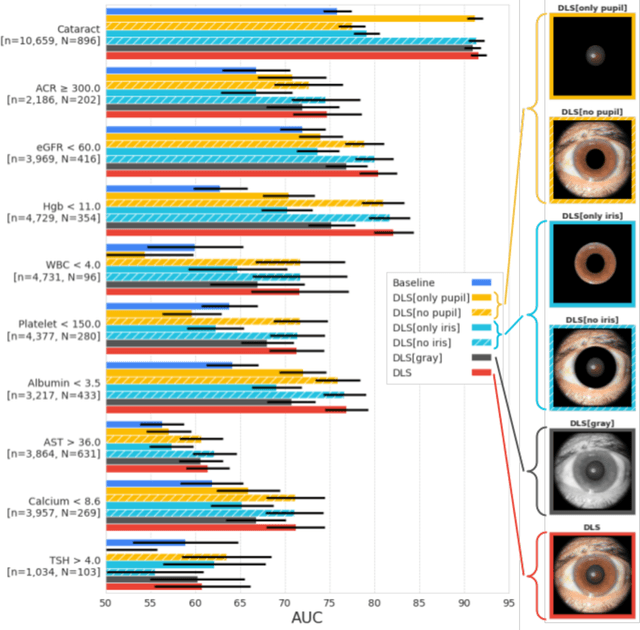
Abstract:External eye photos were recently shown to reveal signs of diabetic retinal disease and elevated HbA1c. In this paper, we evaluate if external eye photos contain information about additional systemic medical conditions. We developed a deep learning system (DLS) that takes external eye photos as input and predicts multiple systemic parameters, such as those related to the liver (albumin, AST); kidney (eGFR estimated using the race-free 2021 CKD-EPI creatinine equation, the urine ACR); bone & mineral (calcium); thyroid (TSH); and blood count (Hgb, WBC, platelets). Development leveraged 151,237 images from 49,015 patients with diabetes undergoing diabetic eye screening in 11 sites across Los Angeles county, CA. Evaluation focused on 9 pre-specified systemic parameters and leveraged 3 validation sets (A, B, C) spanning 28,869 patients with and without diabetes undergoing eye screening in 3 independent sites in Los Angeles County, CA, and the greater Atlanta area, GA. We compared against baseline models incorporating available clinicodemographic variables (e.g. age, sex, race/ethnicity, years with diabetes). Relative to the baseline, the DLS achieved statistically significant superior performance at detecting AST>36, calcium<8.6, eGFR<60, Hgb<11, platelets<150, ACR>=300, and WBC<4 on validation set A (a patient population similar to the development sets), where the AUC of DLS exceeded that of the baseline by 5.2-19.4%. On validation sets B and C, with substantial patient population differences compared to the development sets, the DLS outperformed the baseline for ACR>=300 and Hgb<11 by 7.3-13.2%. Our findings provide further evidence that external eye photos contain important biomarkers of systemic health spanning multiple organ systems. Further work is needed to investigate whether and how these biomarkers can be translated into clinical impact.
Detecting hidden signs of diabetes in external eye photographs
Nov 23, 2020



Abstract:Diabetes-related retinal conditions can be detected by examining the posterior of the eye. By contrast, examining the anterior of the eye can reveal conditions affecting the front of the eye, such as changes to the eyelids, cornea, or crystalline lens. In this work, we studied whether external photographs of the front of the eye can reveal insights into both diabetic retinal diseases and blood glucose control. We developed a deep learning system (DLS) using external eye photographs of 145,832 patients with diabetes from 301 diabetic retinopathy (DR) screening sites in one US state, and evaluated the DLS on three validation sets containing images from 198 sites in 18 other US states. In validation set A (n=27,415 patients, all undilated), the DLS detected poor blood glucose control (HbA1c > 9%) with an area under receiver operating characteristic curve (AUC) of 70.2; moderate-or-worse DR with an AUC of 75.3; diabetic macular edema with an AUC of 78.0; and vision-threatening DR with an AUC of 79.4. For all 4 prediction tasks, the DLS's AUC was higher (p<0.001) than using available self-reported baseline characteristics (age, sex, race/ethnicity, years with diabetes). In terms of positive predictive value, the predicted top 5% of patients had a 67% chance of having HbA1c > 9%, and a 20% chance of having vision threatening diabetic retinopathy. The results generalized to dilated pupils (validation set B, 5,058 patients) and to a different screening service (validation set C, 10,402 patients). Our results indicate that external eye photographs contain information useful for healthcare providers managing patients with diabetes, and may help prioritize patients for in-person screening. Further work is needed to validate these findings on different devices and patient populations (those without diabetes) to evaluate its utility for remote diagnosis and management.
Improving Medical Annotation Quality to Decrease Labeling Burden Using Stratified Noisy Cross-Validation
Sep 22, 2020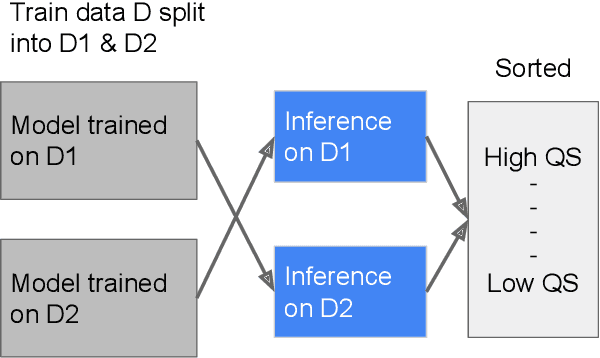

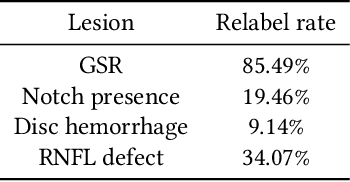
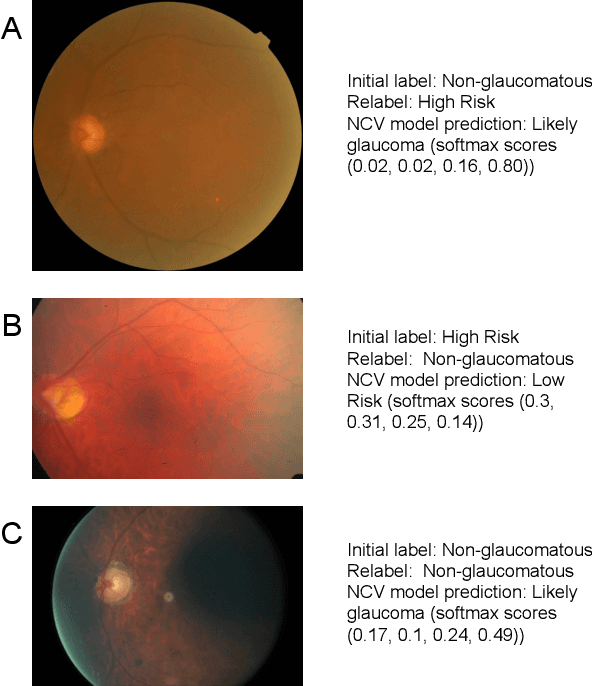
Abstract:As machine learning has become increasingly applied to medical imaging data, noise in training labels has emerged as an important challenge. Variability in diagnosis of medical images is well established; in addition, variability in training and attention to task among medical labelers may exacerbate this issue. Methods for identifying and mitigating the impact of low quality labels have been studied, but are not well characterized in medical imaging tasks. For instance, Noisy Cross-Validation splits the training data into halves, and has been shown to identify low-quality labels in computer vision tasks; but it has not been applied to medical imaging tasks specifically. In this work we introduce Stratified Noisy Cross-Validation (SNCV), an extension of noisy cross validation. SNCV can provide estimates of confidence in model predictions by assigning a quality score to each example; stratify labels to handle class imbalance; and identify likely low-quality labels to analyze the causes. We assess performance of SNCV on diagnosis of glaucoma suspect risk from retinal fundus photographs, a clinically important yet nuanced labeling task. Using training data from a previously-published deep learning model, we compute a continuous quality score (QS) for each training example. We relabel 1,277 low-QS examples using a trained glaucoma specialist; the new labels agree with the SNCV prediction over the initial label >85% of the time, indicating that low-QS examples mostly reflect labeler errors. We then quantify the impact of training with only high-QS labels, showing that strong model performance may be obtained with many fewer examples. By applying the method to randomly sub-sampled training dataset, we show that our method can reduce labelling burden by approximately 50% while achieving model performance non-inferior to using the full dataset on multiple held-out test sets.
Predicting Risk of Developing Diabetic Retinopathy using Deep Learning
Aug 10, 2020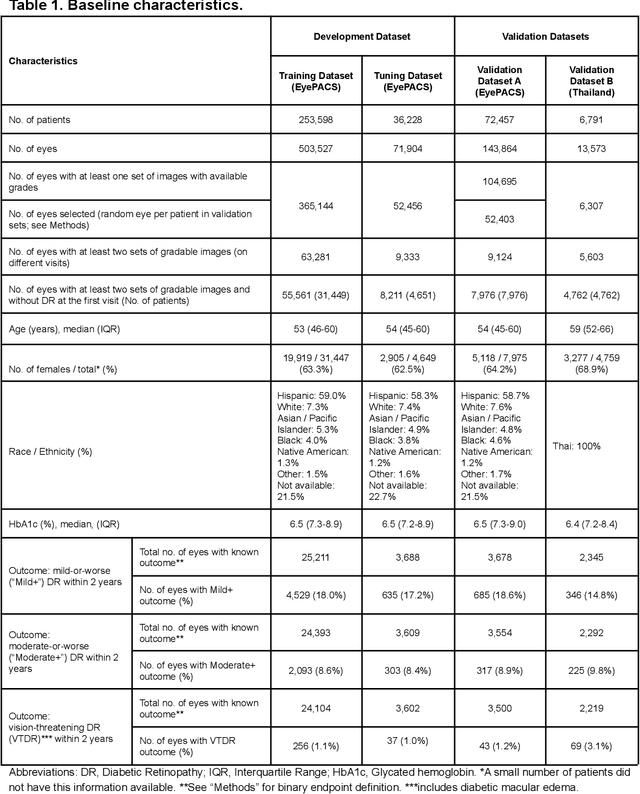

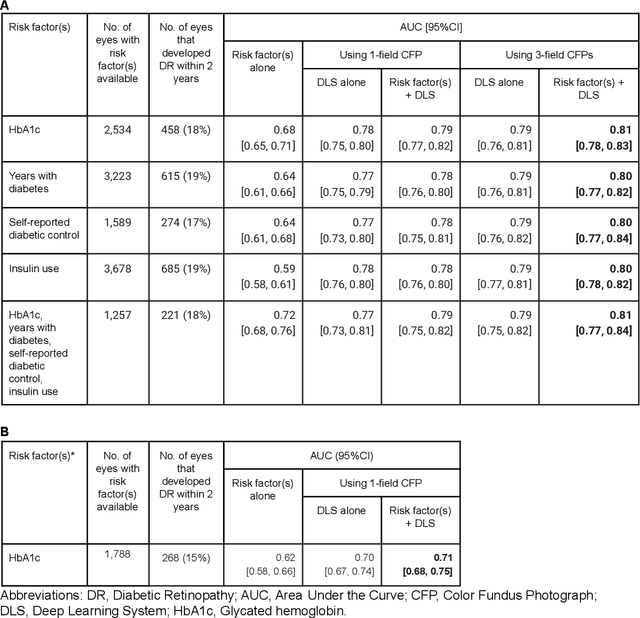

Abstract:Diabetic retinopathy (DR) screening is instrumental in preventing blindness, but faces a scaling challenge as the number of diabetic patients rises. Risk stratification for the development of DR may help optimize screening intervals to reduce costs while improving vision-related outcomes. We created and validated two versions of a deep learning system (DLS) to predict the development of mild-or-worse ("Mild+") DR in diabetic patients undergoing DR screening. The two versions used either three-fields or a single field of color fundus photographs (CFPs) as input. The training set was derived from 575,431 eyes, of which 28,899 had known 2-year outcome, and the remaining were used to augment the training process via multi-task learning. Validation was performed on both an internal validation set (set A; 7,976 eyes; 3,678 with known outcome) and an external validation set (set B; 4,762 eyes; 2,345 with known outcome). For predicting 2-year development of DR, the 3-field DLS had an area under the receiver operating characteristic curve (AUC) of 0.79 (95%CI, 0.78-0.81) on validation set A. On validation set B (which contained only a single field), the 1-field DLS's AUC was 0.70 (95%CI, 0.67-0.74). The DLS was prognostic even after adjusting for available risk factors (p<0.001). When added to the risk factors, the 3-field DLS improved the AUC from 0.72 (95%CI, 0.68-0.76) to 0.81 (95%CI, 0.77-0.84) in validation set A, and the 1-field DLS improved the AUC from 0.62 (95%CI, 0.58-0.66) to 0.71 (95%CI, 0.68-0.75) in validation set B. The DLSs in this study identified prognostic information for DR development from CFPs. This information is independent of and more informative than the available risk factors.
Detecting Anemia from Retinal Fundus Images
Apr 12, 2019



Abstract:Despite its high prevalence, anemia is often undetected due to the invasiveness and cost of screening and diagnostic tests. Though some non-invasive approaches have been developed, they are less accurate than invasive methods, resulting in an unmet need for more accurate non-invasive methods. Here, we show that deep learning-based algorithms can detect anemia and quantify several related blood measurements using retinal fundus images both in isolation and in combination with basic metadata such as patient demographics. On a validation dataset of 11,388 patients from the UK Biobank, our algorithms achieved a mean absolute error of 0.63 g/dL (95% confidence interval (CI) 0.62-0.64) in quantifying hemoglobin concentration and an area under receiver operating characteristic curve (AUC) of 0.88 (95% CI 0.86-0.89) in detecting anemia. This work shows the potential of automated non-invasive anemia screening based on fundus images, particularly in diabetic patients, who may have regular retinal imaging and are at increased risk of further morbidity and mortality from anemia.
Predicting Progression of Age-related Macular Degeneration from Fundus Images using Deep Learning
Apr 10, 2019



Abstract:Background: Patients with neovascular age-related macular degeneration (AMD) can avoid vision loss via certain therapy. However, methods to predict the progression to neovascular age-related macular degeneration (nvAMD) are lacking. Purpose: To develop and validate a deep learning (DL) algorithm to predict 1-year progression of eyes with no, early, or intermediate AMD to nvAMD, using color fundus photographs (CFP). Design: Development and validation of a DL algorithm. Methods: We trained a DL algorithm to predict 1-year progression to nvAMD, and used 10-fold cross-validation to evaluate this approach on two groups of eyes in the Age-Related Eye Disease Study (AREDS): none/early/intermediate AMD, and intermediate AMD (iAMD) only. We compared the DL algorithm to the manually graded 4-category and 9-step scales in the AREDS dataset. Main outcome measures: Performance of the DL algorithm was evaluated using the sensitivity at 80% specificity for progression to nvAMD. Results: The DL algorithm's sensitivity for predicting progression to nvAMD from none/early/iAMD (78+/-6%) was higher than manual grades from the 9-step scale (67+/-8%) or the 4-category scale (48+/-3%). For predicting progression specifically from iAMD, the DL algorithm's sensitivity (57+/-6%) was also higher compared to the 9-step grades (36+/-8%) and the 4-category grades (20+/-0%). Conclusions: Our DL algorithm performed better in predicting progression to nvAMD than manual grades. Future investigations are required to test the application of this DL algorithm in a real-world clinical setting.
Deep Learning to Assess Glaucoma Risk and Associated Features in Fundus Images
Dec 21, 2018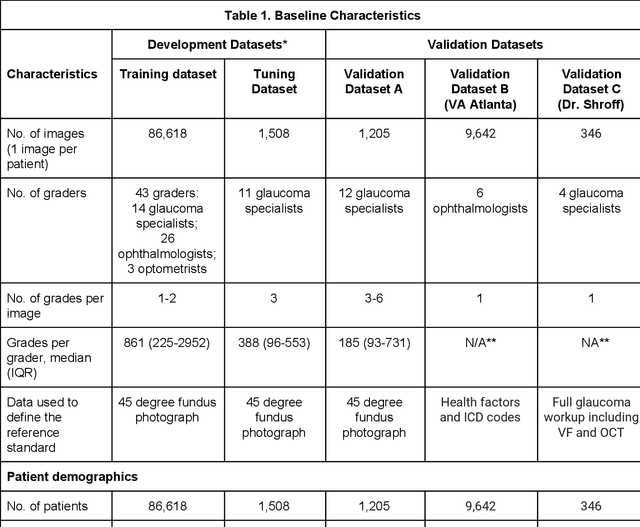
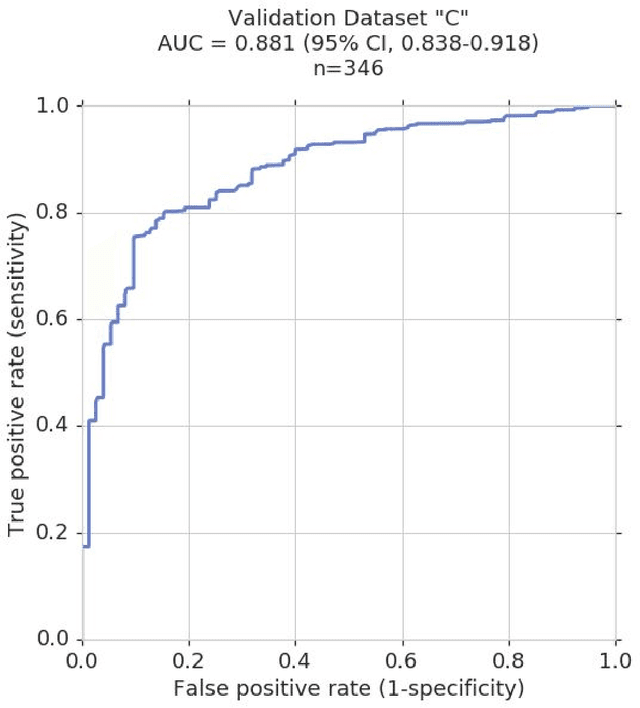
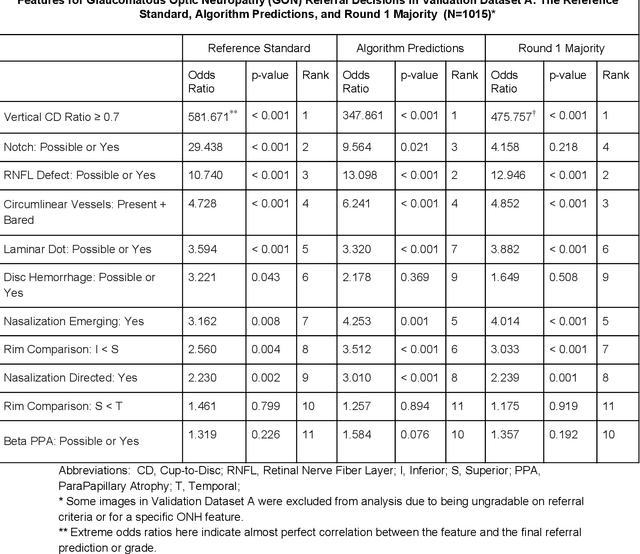
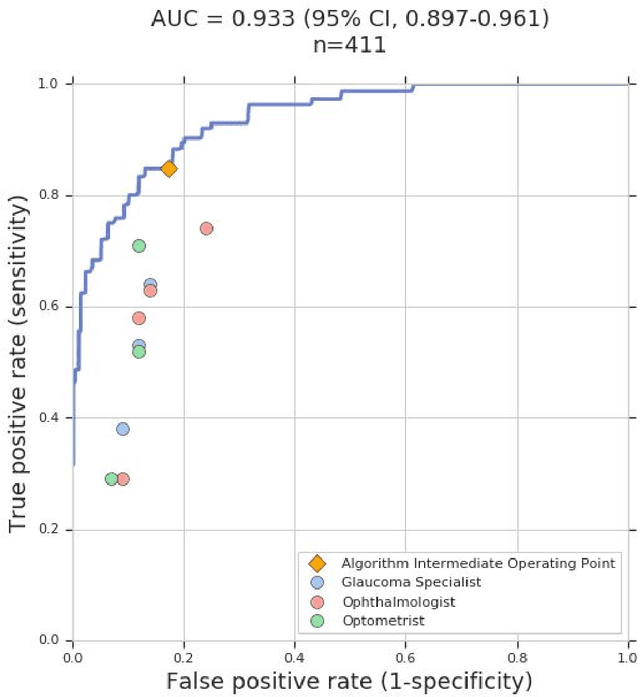
Abstract:Glaucoma is the leading cause of preventable, irreversible blindness world-wide. The disease can remain asymptomatic until severe, and an estimated 50%-90% of people with glaucoma remain undiagnosed. Thus, glaucoma screening is recommended for early detection and treatment. A cost-effective tool to detect glaucoma could expand healthcare access to a much larger patient population, but such a tool is currently unavailable. We trained a deep learning (DL) algorithm using a retrospective dataset of 58,033 images, assessed for gradability, glaucomatous optic nerve head (ONH) features, and referable glaucoma risk. The resultant algorithm was validated using 2 separate datasets. For referable glaucoma risk, the algorithm had an AUC of 0.940 (95%CI, 0.922-0.955) in validation dataset "A" (1,205 images, 1 image/patient; 19% referable where images were adjudicated by panels of fellowship-trained glaucoma specialists) and 0.858 (95% CI, 0.836-0.878) in validation dataset "B" (17,593 images from 9,643 patients; 9.2% referable where images were from the Atlanta Veterans Affairs Eye Clinic diabetic teleretinal screening program using clinical referral decisions as the reference standard). Additionally, we found that the presence of vertical cup-to-disc ratio >= 0.7, neuroretinal rim notching, retinal nerve fiber layer defect, and bared circumlinear vessels contributed most to referable glaucoma risk assessment by both glaucoma specialists and the algorithm. Algorithm AUCs ranged between 0.608-0.977 for glaucomatous ONH features. The DL algorithm was significantly more sensitive than 6 of 10 graders, including 2 of 3 glaucoma specialists, with comparable or higher specificity relative to all graders. A DL algorithm trained on fundus images alone can detect referable glaucoma risk with higher sensitivity and comparable specificity to eye care providers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge