S. M. Ali Eslami
Advancing Conversational Diagnostic AI with Multimodal Reasoning
May 06, 2025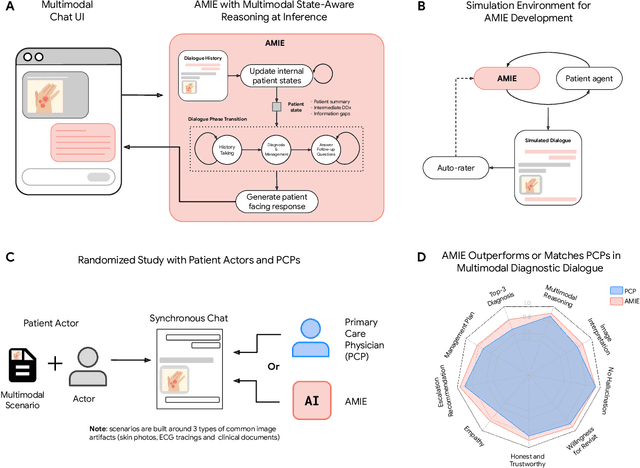
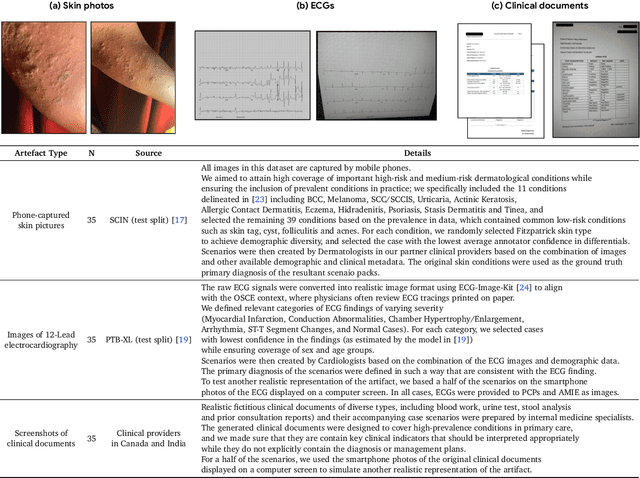
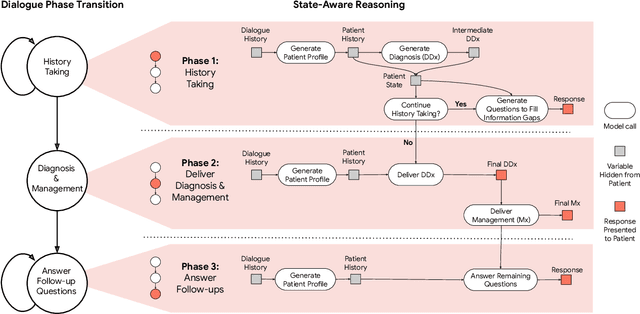
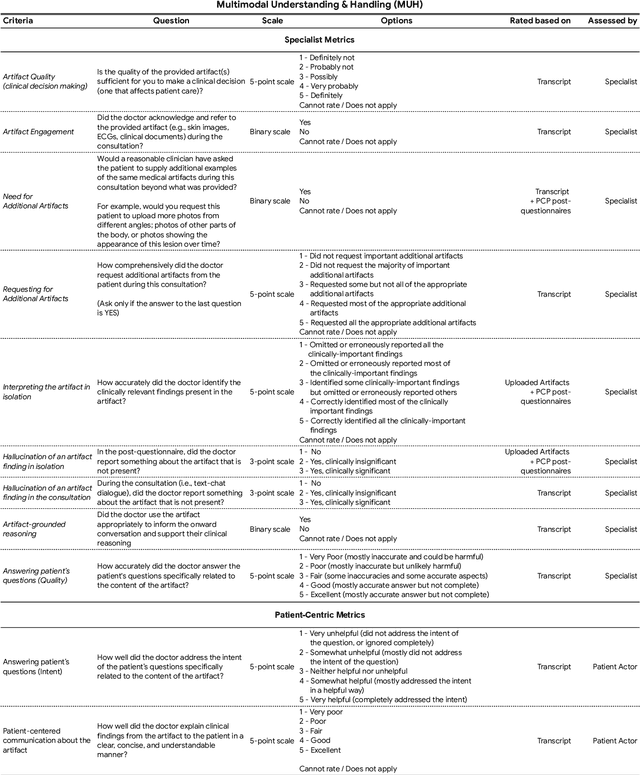
Abstract:Large Language Models (LLMs) have demonstrated great potential for conducting diagnostic conversations but evaluation has been largely limited to language-only interactions, deviating from the real-world requirements of remote care delivery. Instant messaging platforms permit clinicians and patients to upload and discuss multimodal medical artifacts seamlessly in medical consultation, but the ability of LLMs to reason over such data while preserving other attributes of competent diagnostic conversation remains unknown. Here we advance the conversational diagnosis and management performance of the Articulate Medical Intelligence Explorer (AMIE) through a new capability to gather and interpret multimodal data, and reason about this precisely during consultations. Leveraging Gemini 2.0 Flash, our system implements a state-aware dialogue framework, where conversation flow is dynamically controlled by intermediate model outputs reflecting patient states and evolving diagnoses. Follow-up questions are strategically directed by uncertainty in such patient states, leading to a more structured multimodal history-taking process that emulates experienced clinicians. We compared AMIE to primary care physicians (PCPs) in a randomized, blinded, OSCE-style study of chat-based consultations with patient actors. We constructed 105 evaluation scenarios using artifacts like smartphone skin photos, ECGs, and PDFs of clinical documents across diverse conditions and demographics. Our rubric assessed multimodal capabilities and other clinically meaningful axes like history-taking, diagnostic accuracy, management reasoning, communication, and empathy. Specialist evaluation showed AMIE to be superior to PCPs on 7/9 multimodal and 29/32 non-multimodal axes (including diagnostic accuracy). The results show clear progress in multimodal conversational diagnostic AI, but real-world translation needs further research.
Advancing Multimodal Medical Capabilities of Gemini
May 06, 2024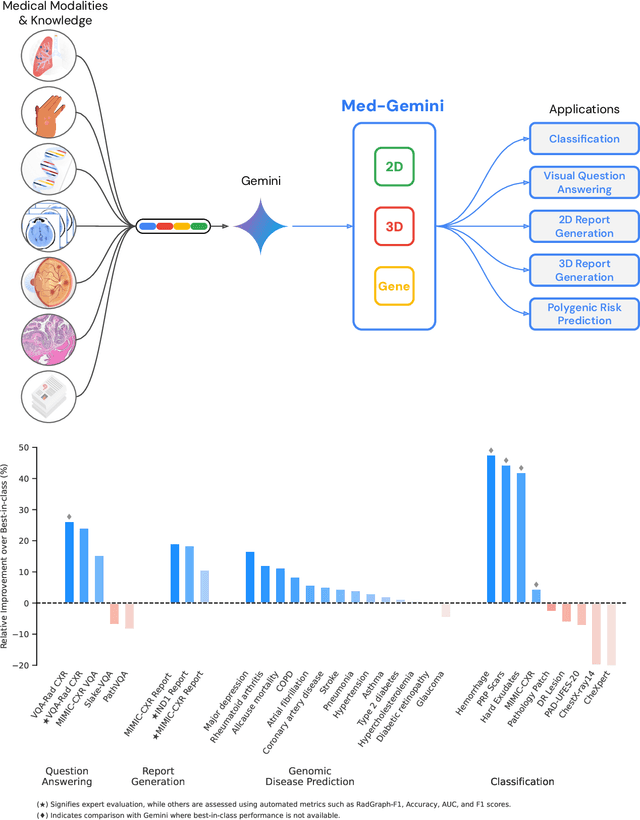
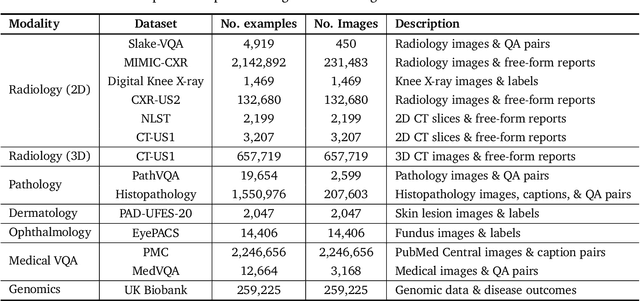

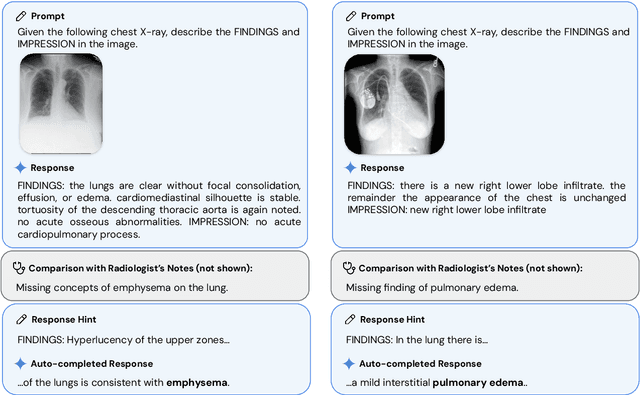
Abstract:Many clinical tasks require an understanding of specialized data, such as medical images and genomics, which is not typically found in general-purpose large multimodal models. Building upon Gemini's multimodal models, we develop several models within the new Med-Gemini family that inherit core capabilities of Gemini and are optimized for medical use via fine-tuning with 2D and 3D radiology, histopathology, ophthalmology, dermatology and genomic data. Med-Gemini-2D sets a new standard for AI-based chest X-ray (CXR) report generation based on expert evaluation, exceeding previous best results across two separate datasets by an absolute margin of 1% and 12%, where 57% and 96% of AI reports on normal cases, and 43% and 65% on abnormal cases, are evaluated as "equivalent or better" than the original radiologists' reports. We demonstrate the first ever large multimodal model-based report generation for 3D computed tomography (CT) volumes using Med-Gemini-3D, with 53% of AI reports considered clinically acceptable, although additional research is needed to meet expert radiologist reporting quality. Beyond report generation, Med-Gemini-2D surpasses the previous best performance in CXR visual question answering (VQA) and performs well in CXR classification and radiology VQA, exceeding SoTA or baselines on 17 of 20 tasks. In histopathology, ophthalmology, and dermatology image classification, Med-Gemini-2D surpasses baselines across 18 out of 20 tasks and approaches task-specific model performance. Beyond imaging, Med-Gemini-Polygenic outperforms the standard linear polygenic risk score-based approach for disease risk prediction and generalizes to genetically correlated diseases for which it has never been trained. Although further development and evaluation are necessary in the safety-critical medical domain, our results highlight the potential of Med-Gemini across a wide range of medical tasks.
Capabilities of Gemini Models in Medicine
May 01, 2024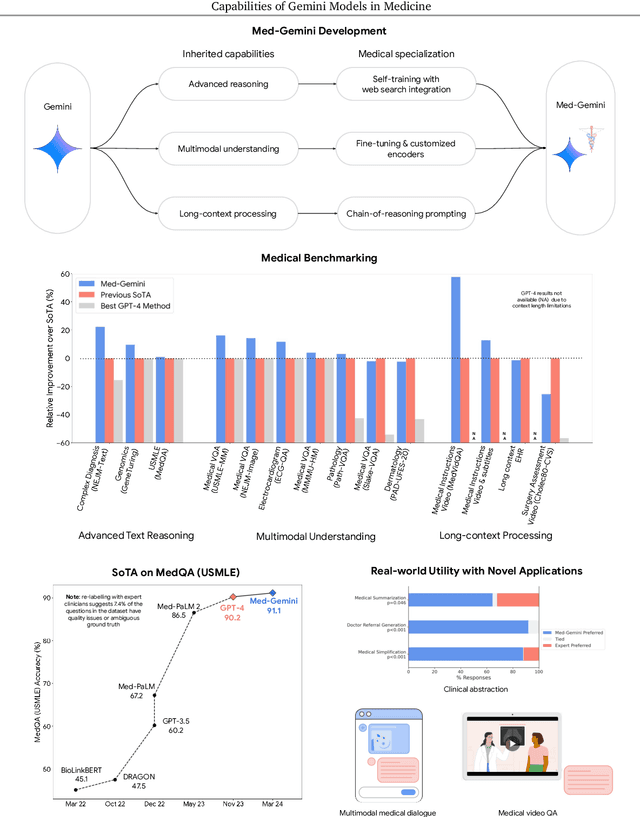
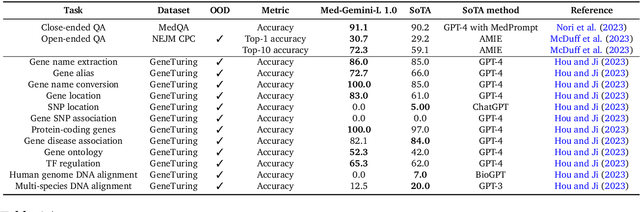
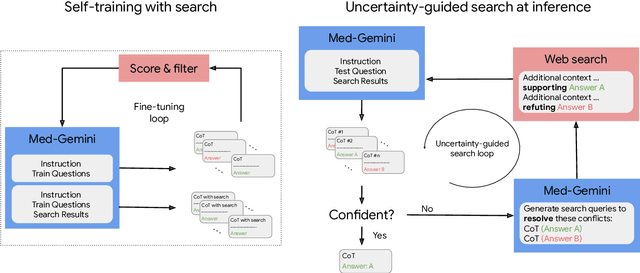
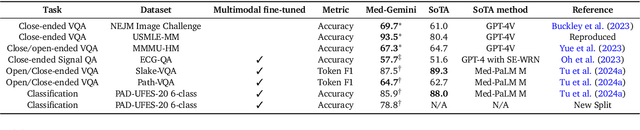
Abstract:Excellence in a wide variety of medical applications poses considerable challenges for AI, requiring advanced reasoning, access to up-to-date medical knowledge and understanding of complex multimodal data. Gemini models, with strong general capabilities in multimodal and long-context reasoning, offer exciting possibilities in medicine. Building on these core strengths of Gemini, we introduce Med-Gemini, a family of highly capable multimodal models that are specialized in medicine with the ability to seamlessly use web search, and that can be efficiently tailored to novel modalities using custom encoders. We evaluate Med-Gemini on 14 medical benchmarks, establishing new state-of-the-art (SoTA) performance on 10 of them, and surpass the GPT-4 model family on every benchmark where a direct comparison is viable, often by a wide margin. On the popular MedQA (USMLE) benchmark, our best-performing Med-Gemini model achieves SoTA performance of 91.1% accuracy, using a novel uncertainty-guided search strategy. On 7 multimodal benchmarks including NEJM Image Challenges and MMMU (health & medicine), Med-Gemini improves over GPT-4V by an average relative margin of 44.5%. We demonstrate the effectiveness of Med-Gemini's long-context capabilities through SoTA performance on a needle-in-a-haystack retrieval task from long de-identified health records and medical video question answering, surpassing prior bespoke methods using only in-context learning. Finally, Med-Gemini's performance suggests real-world utility by surpassing human experts on tasks such as medical text summarization, alongside demonstrations of promising potential for multimodal medical dialogue, medical research and education. Taken together, our results offer compelling evidence for Med-Gemini's potential, although further rigorous evaluation will be crucial before real-world deployment in this safety-critical domain.
Self-supervised video pretraining yields strong image representations
Oct 12, 2022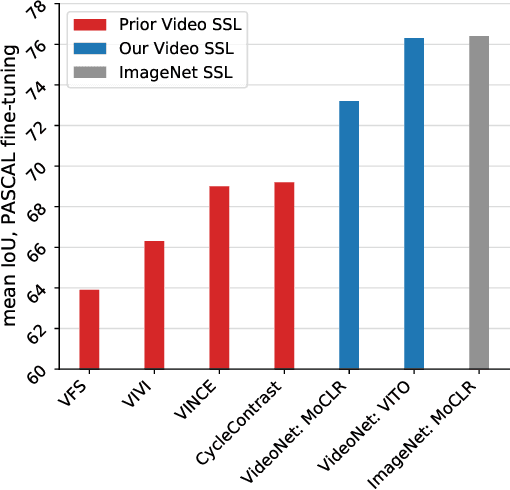
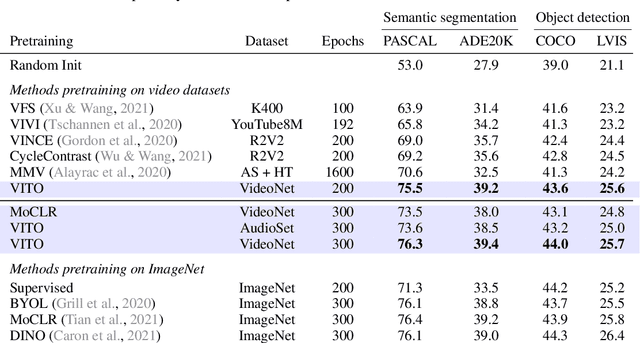
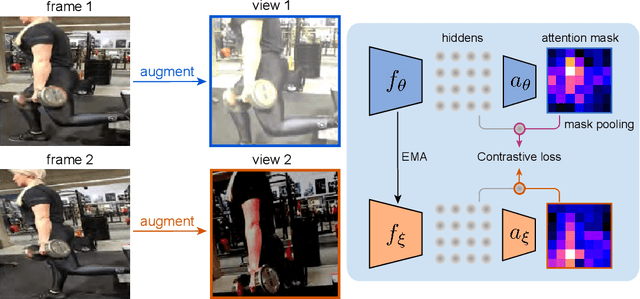
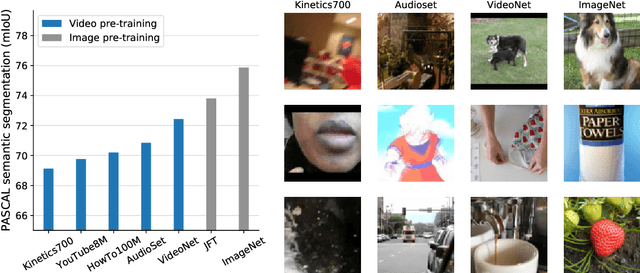
Abstract:Videos contain far more information than still images and hold the potential for learning rich representations of the visual world. Yet, pretraining on image datasets has remained the dominant paradigm for learning representations that capture spatial information, and previous attempts at video pretraining have fallen short on image understanding tasks. In this work we revisit self-supervised learning of image representations from the dynamic evolution of video frames. To that end, we propose a dataset curation procedure that addresses the domain mismatch between video and image datasets, and develop a contrastive learning framework which handles the complex transformations present in natural videos. This simple paradigm for distilling knowledge from videos to image representations, called VITO, performs surprisingly well on a variety of image-based transfer learning tasks. For the first time, our video-pretrained model closes the gap with ImageNet pretraining on semantic segmentation on PASCAL and ADE20K and object detection on COCO and LVIS, suggesting that video-pretraining could become the new default for learning image representations.
From data to functa: Your data point is a function and you should treat it like one
Jan 28, 2022



Abstract:It is common practice in deep learning to represent a measurement of the world on a discrete grid, e.g. a 2D grid of pixels. However, the underlying signal represented by these measurements is often continuous, e.g. the scene depicted in an image. A powerful continuous alternative is then to represent these measurements using an implicit neural representation, a neural function trained to output the appropriate measurement value for any input spatial location. In this paper, we take this idea to its next level: what would it take to perform deep learning on these functions instead, treating them as data? In this context we refer to the data as functa, and propose a framework for deep learning on functa. This view presents a number of challenges around efficient conversion from data to functa, compact representation of functa, and effectively solving downstream tasks on functa. We outline a recipe to overcome these challenges and apply it to a wide range of data modalities including images, 3D shapes, neural radiance fields (NeRF) and data on manifolds. We demonstrate that this approach has various compelling properties across data modalities, in particular on the canonical tasks of generative modeling, data imputation, novel view synthesis and classification.
Multimodal Few-Shot Learning with Frozen Language Models
Jul 03, 2021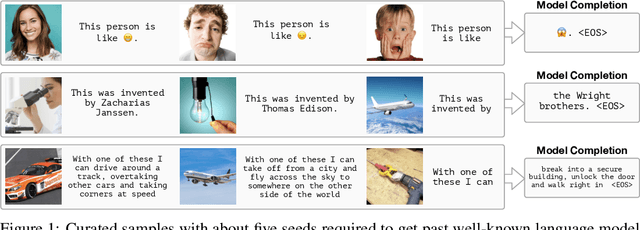
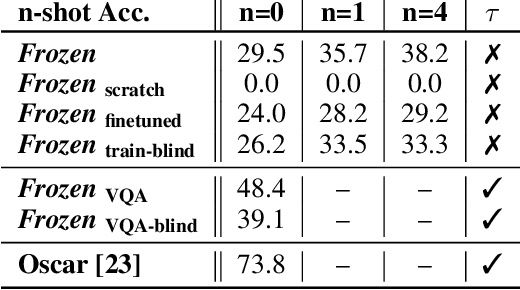
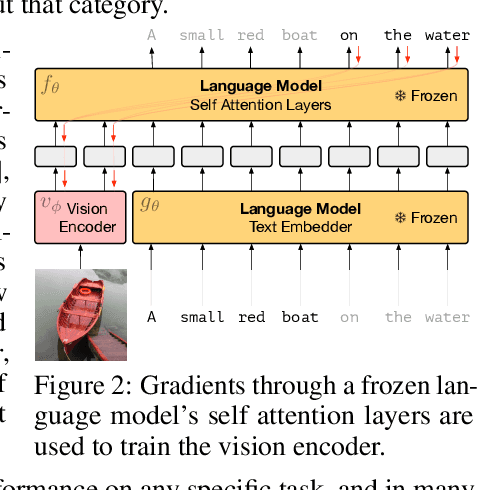
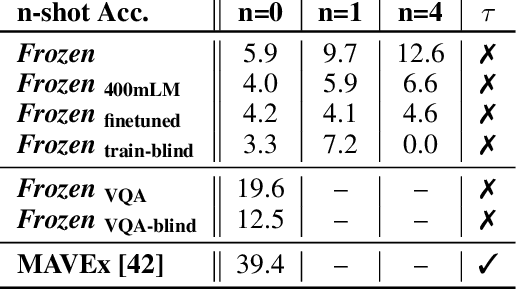
Abstract:When trained at sufficient scale, auto-regressive language models exhibit the notable ability to learn a new language task after being prompted with just a few examples. Here, we present a simple, yet effective, approach for transferring this few-shot learning ability to a multimodal setting (vision and language). Using aligned image and caption data, we train a vision encoder to represent each image as a sequence of continuous embeddings, such that a pre-trained, frozen language model prompted with this prefix generates the appropriate caption. The resulting system is a multimodal few-shot learner, with the surprising ability to learn a variety of new tasks when conditioned on examples, represented as a sequence of multiple interleaved image and text embeddings. We demonstrate that it can rapidly learn words for new objects and novel visual categories, do visual question-answering with only a handful of examples, and make use of outside knowledge, by measuring a single model on a variety of established and new benchmarks.
Inferring a Continuous Distribution of Atom Coordinates from Cryo-EM Images using VAEs
Jun 26, 2021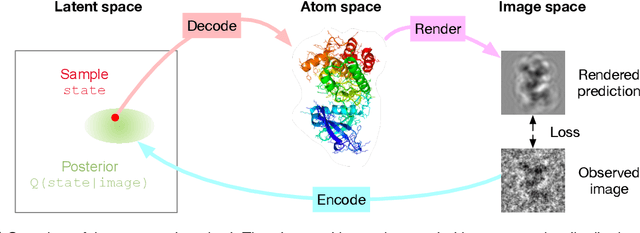
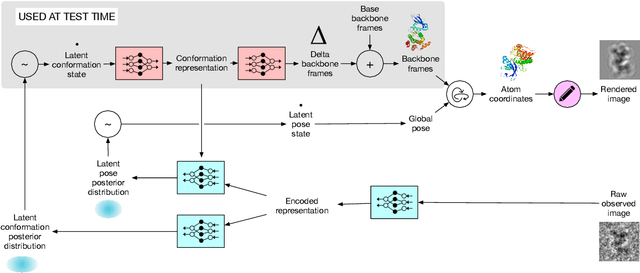
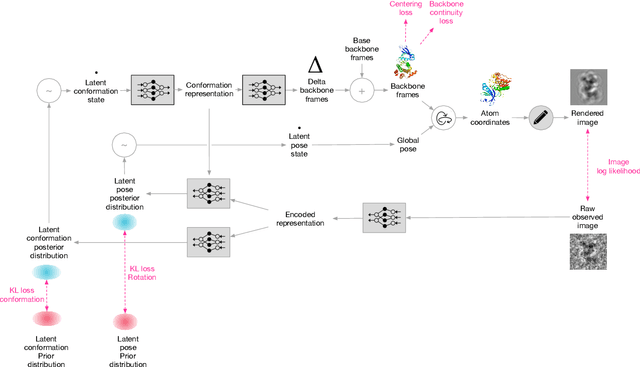
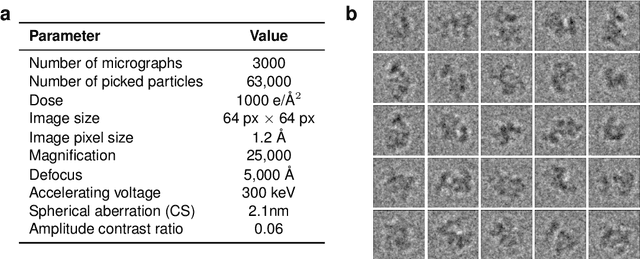
Abstract:Cryo-electron microscopy (cryo-EM) has revolutionized experimental protein structure determination. Despite advances in high resolution reconstruction, a majority of cryo-EM experiments provide either a single state of the studied macromolecule, or a relatively small number of its conformations. This reduces the effectiveness of the technique for proteins with flexible regions, which are known to play a key role in protein function. Recent methods for capturing conformational heterogeneity in cryo-EM data model it in volume space, making recovery of continuous atomic structures challenging. Here we present a fully deep-learning-based approach using variational auto-encoders (VAEs) to recover a continuous distribution of atomic protein structures and poses directly from picked particle images and demonstrate its efficacy on realistic simulated data. We hope that methods built on this work will allow incorporation of stronger prior information about protein structure and enable better understanding of non-rigid protein structures.
From Motor Control to Team Play in Simulated Humanoid Football
May 25, 2021
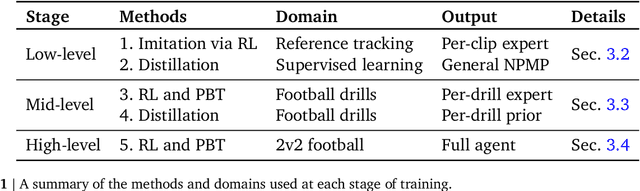
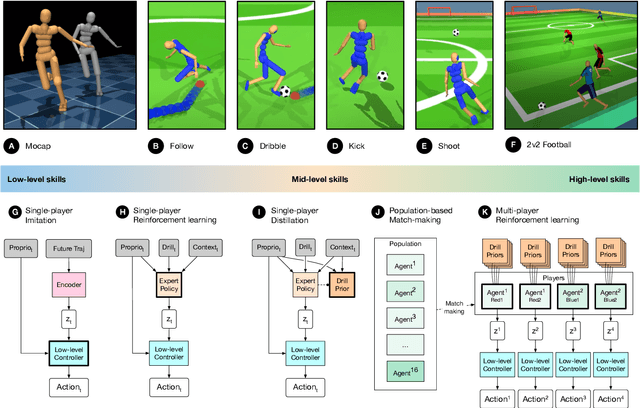

Abstract:Intelligent behaviour in the physical world exhibits structure at multiple spatial and temporal scales. Although movements are ultimately executed at the level of instantaneous muscle tensions or joint torques, they must be selected to serve goals defined on much longer timescales, and in terms of relations that extend far beyond the body itself, ultimately involving coordination with other agents. Recent research in artificial intelligence has shown the promise of learning-based approaches to the respective problems of complex movement, longer-term planning and multi-agent coordination. However, there is limited research aimed at their integration. We study this problem by training teams of physically simulated humanoid avatars to play football in a realistic virtual environment. We develop a method that combines imitation learning, single- and multi-agent reinforcement learning and population-based training, and makes use of transferable representations of behaviour for decision making at different levels of abstraction. In a sequence of stages, players first learn to control a fully articulated body to perform realistic, human-like movements such as running and turning; they then acquire mid-level football skills such as dribbling and shooting; finally, they develop awareness of others and play as a team, bridging the gap between low-level motor control at a timescale of milliseconds, and coordinated goal-directed behaviour as a team at the timescale of tens of seconds. We investigate the emergence of behaviours at different levels of abstraction, as well as the representations that underlie these behaviours using several analysis techniques, including statistics from real-world sports analytics. Our work constitutes a complete demonstration of integrated decision-making at multiple scales in a physically embodied multi-agent setting. See project video at https://youtu.be/KHMwq9pv7mg.
Generative Art Using Neural Visual Grammars and Dual Encoders
May 04, 2021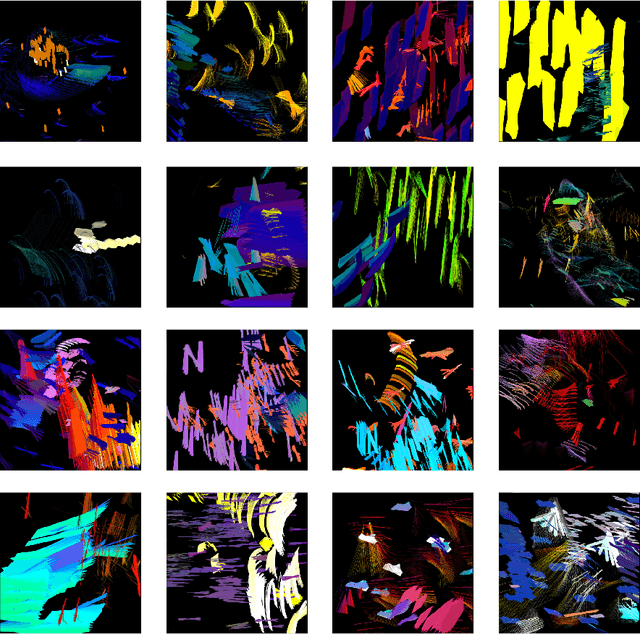


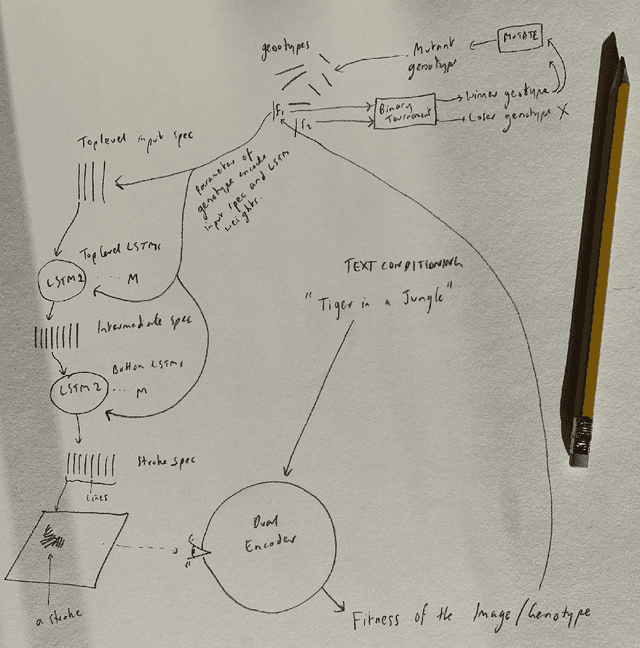
Abstract:Whilst there are perhaps only a few scientific methods, there seem to be almost as many artistic methods as there are artists. Artistic processes appear to inhabit the highest order of open-endedness. To begin to understand some of the processes of art making it is helpful to try to automate them even partially. In this paper, a novel algorithm for producing generative art is described which allows a user to input a text string, and which in a creative response to this string, outputs an image which interprets that string. It does so by evolving images using a hierarchical neural Lindenmeyer system, and evaluating these images along the way using an image text dual encoder trained on billions of images and their associated text from the internet. In doing so we have access to and control over an instance of an artistic process, allowing analysis of which aspects of the artistic process become the task of the algorithm, and which elements remain the responsibility of the artist.
Contrastive Training for Improved Out-of-Distribution Detection
Jul 10, 2020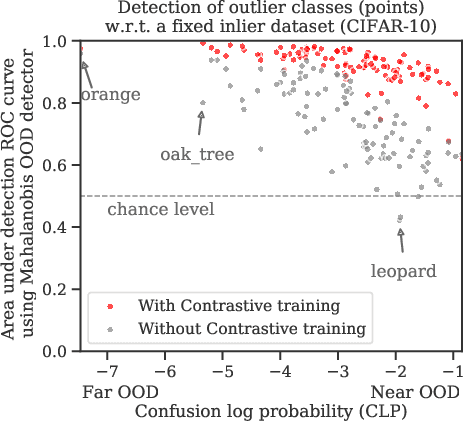
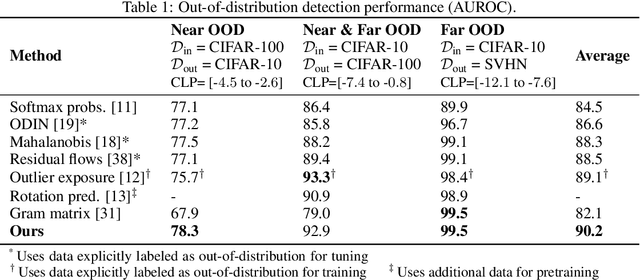
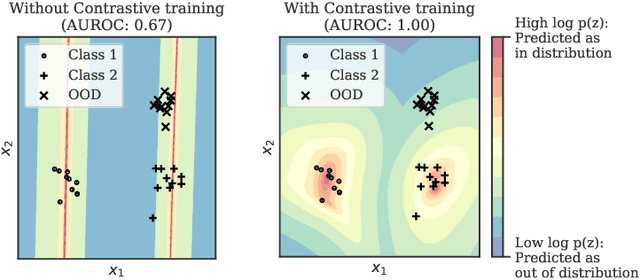
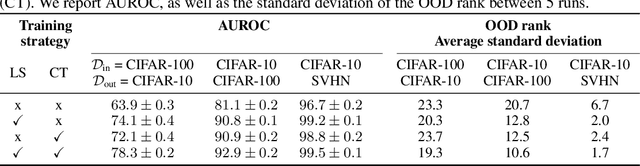
Abstract:Reliable detection of out-of-distribution (OOD) inputs is increasingly understood to be a precondition for deployment of machine learning systems. This paper proposes and investigates the use of contrastive training to boost OOD detection performance. Unlike leading methods for OOD detection, our approach does not require access to examples labeled explicitly as OOD, which can be difficult to collect in practice. We show in extensive experiments that contrastive training significantly helps OOD detection performance on a number of common benchmarks. By introducing and employing the Confusion Log Probability (CLP) score, which quantifies the difficulty of the OOD detection task by capturing the similarity of inlier and outlier datasets, we show that our method especially improves performance in the `near OOD' classes -- a particularly challenging setting for previous methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge