John Jumper
Accelerating Large Language Model Decoding with Speculative Sampling
Feb 02, 2023


Abstract:We present speculative sampling, an algorithm for accelerating transformer decoding by enabling the generation of multiple tokens from each transformer call. Our algorithm relies on the observation that the latency of parallel scoring of short continuations, generated by a faster but less powerful draft model, is comparable to that of sampling a single token from the larger target model. This is combined with a novel modified rejection sampling scheme which preserves the distribution of the target model within hardware numerics. We benchmark speculative sampling with Chinchilla, a 70 billion parameter language model, achieving a 2-2.5x decoding speedup in a distributed setup, without compromising the sample quality or making modifications to the model itself.
Inferring a Continuous Distribution of Atom Coordinates from Cryo-EM Images using VAEs
Jun 26, 2021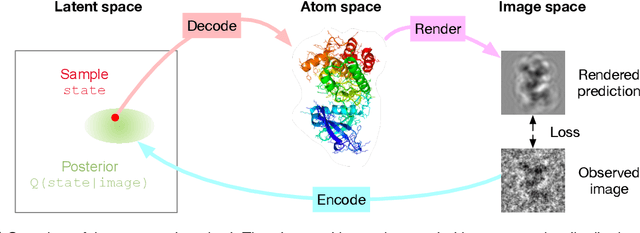
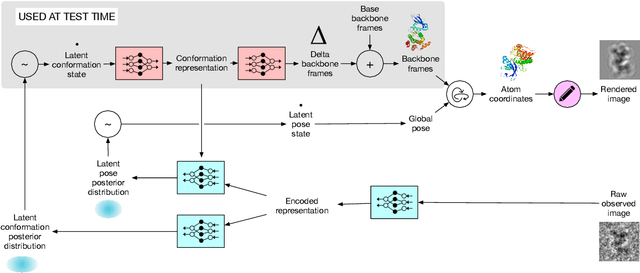
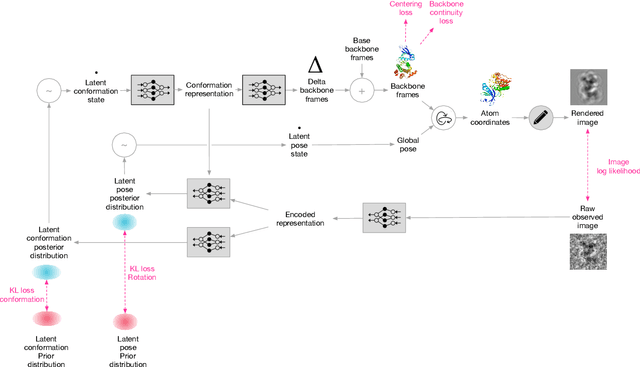
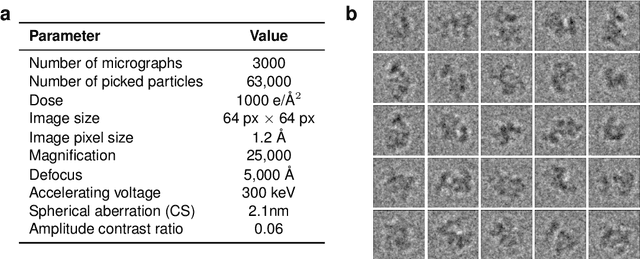
Abstract:Cryo-electron microscopy (cryo-EM) has revolutionized experimental protein structure determination. Despite advances in high resolution reconstruction, a majority of cryo-EM experiments provide either a single state of the studied macromolecule, or a relatively small number of its conformations. This reduces the effectiveness of the technique for proteins with flexible regions, which are known to play a key role in protein function. Recent methods for capturing conformational heterogeneity in cryo-EM data model it in volume space, making recovery of continuous atomic structures challenging. Here we present a fully deep-learning-based approach using variational auto-encoders (VAEs) to recover a continuous distribution of atomic protein structures and poses directly from picked particle images and demonstrate its efficacy on realistic simulated data. We hope that methods built on this work will allow incorporation of stronger prior information about protein structure and enable better understanding of non-rigid protein structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge