Andrew W. Senior
A scalable and real-time neural decoder for topological quantum codes
Dec 08, 2025Abstract:Fault-tolerant quantum computing will require error rates far below those achievable with physical qubits. Quantum error correction (QEC) bridges this gap, but depends on decoders being simultaneously fast, accurate, and scalable. This combination of requirements has not yet been met by a machine-learning decoder, nor by any decoder for promising resource-efficient codes such as the colour code. Here we introduce AlphaQubit 2, a neural-network decoder that achieves near-optimal logical error rates for both surface and colour codes at large scales under realistic noise. For the colour code, it is orders of magnitude faster than other high-accuracy decoders. For the surface code, we demonstrate real-time decoding faster than 1 microsecond per cycle up to distance 11 on current commercial accelerators with better accuracy than leading real-time decoders. These results support the practical application of a wider class of promising QEC codes, and establish a credible path towards high-accuracy, real-time neural decoding at the scales required for fault-tolerant quantum computation.
Inferring a Continuous Distribution of Atom Coordinates from Cryo-EM Images using VAEs
Jun 26, 2021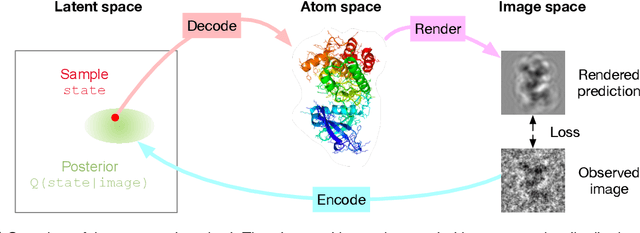
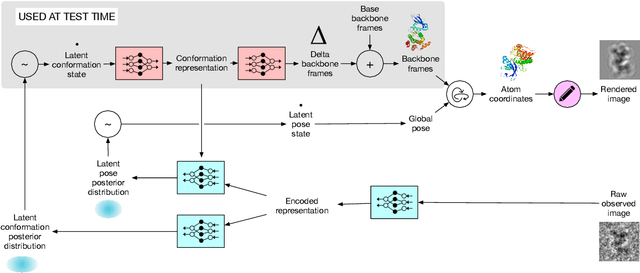
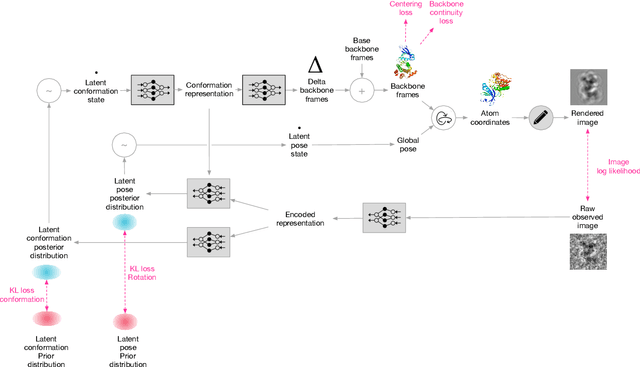
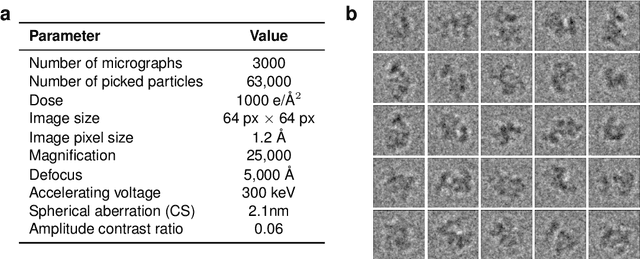
Abstract:Cryo-electron microscopy (cryo-EM) has revolutionized experimental protein structure determination. Despite advances in high resolution reconstruction, a majority of cryo-EM experiments provide either a single state of the studied macromolecule, or a relatively small number of its conformations. This reduces the effectiveness of the technique for proteins with flexible regions, which are known to play a key role in protein function. Recent methods for capturing conformational heterogeneity in cryo-EM data model it in volume space, making recovery of continuous atomic structures challenging. Here we present a fully deep-learning-based approach using variational auto-encoders (VAEs) to recover a continuous distribution of atomic protein structures and poses directly from picked particle images and demonstrate its efficacy on realistic simulated data. We hope that methods built on this work will allow incorporation of stronger prior information about protein structure and enable better understanding of non-rigid protein structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge