Thomas Edlich
A scalable and real-time neural decoder for topological quantum codes
Dec 08, 2025Abstract:Fault-tolerant quantum computing will require error rates far below those achievable with physical qubits. Quantum error correction (QEC) bridges this gap, but depends on decoders being simultaneously fast, accurate, and scalable. This combination of requirements has not yet been met by a machine-learning decoder, nor by any decoder for promising resource-efficient codes such as the colour code. Here we introduce AlphaQubit 2, a neural-network decoder that achieves near-optimal logical error rates for both surface and colour codes at large scales under realistic noise. For the colour code, it is orders of magnitude faster than other high-accuracy decoders. For the surface code, we demonstrate real-time decoding faster than 1 microsecond per cycle up to distance 11 on current commercial accelerators with better accuracy than leading real-time decoders. These results support the practical application of a wider class of promising QEC codes, and establish a credible path towards high-accuracy, real-time neural decoding at the scales required for fault-tolerant quantum computation.
The unknotting number, hard unknot diagrams, and reinforcement learning
Sep 13, 2024



Abstract:We have developed a reinforcement learning agent that often finds a minimal sequence of unknotting crossing changes for a knot diagram with up to 200 crossings, hence giving an upper bound on the unknotting number. We have used this to determine the unknotting number of 57k knots. We took diagrams of connected sums of such knots with oppositely signed signatures, where the summands were overlaid. The agent has found examples where several of the crossing changes in an unknotting collection of crossings result in hyperbolic knots. Based on this, we have shown that, given knots $K$ and $K'$ that satisfy some mild assumptions, there is a diagram of their connected sum and $u(K) + u(K')$ unknotting crossings such that changing any one of them results in a prime knot. As a by-product, we have obtained a dataset of 2.6 million distinct hard unknot diagrams; most of them under 35 crossings. Assuming the additivity of the unknotting number, we have determined the unknotting number of 43 at most 12-crossing knots for which the unknotting number is unknown.
Learning to Decode the Surface Code with a Recurrent, Transformer-Based Neural Network
Oct 09, 2023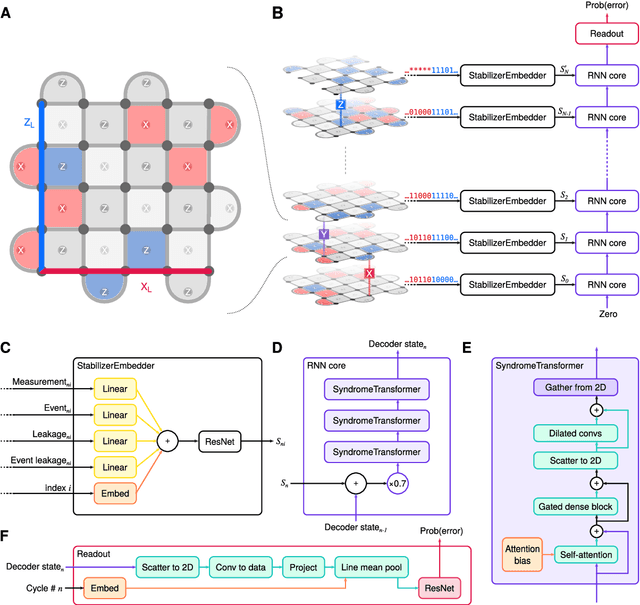
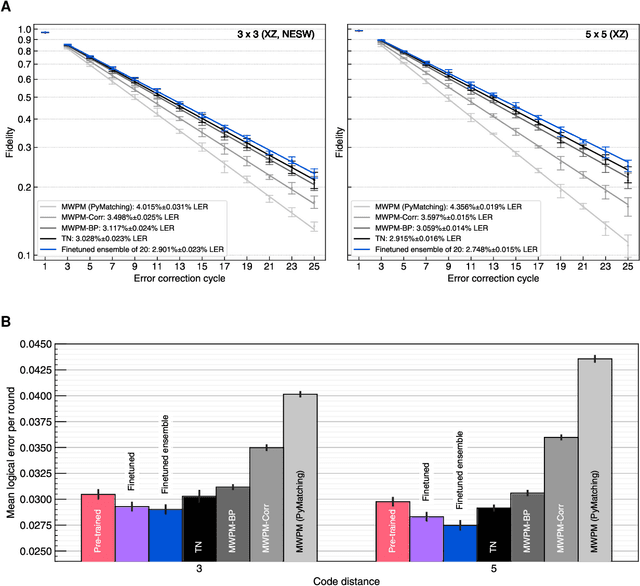
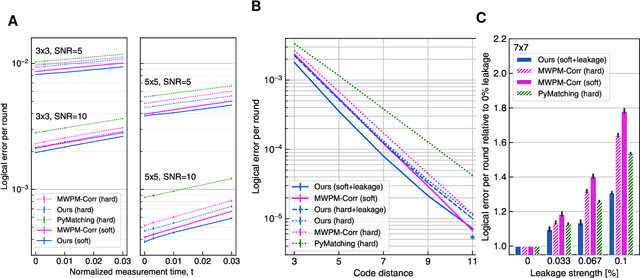
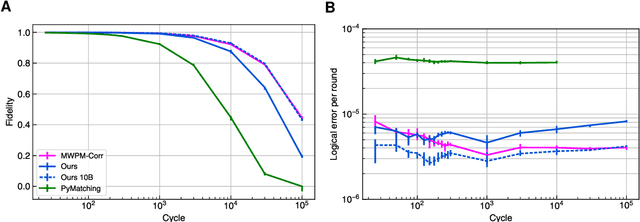
Abstract:Quantum error-correction is a prerequisite for reliable quantum computation. Towards this goal, we present a recurrent, transformer-based neural network which learns to decode the surface code, the leading quantum error-correction code. Our decoder outperforms state-of-the-art algorithmic decoders on real-world data from Google's Sycamore quantum processor for distance 3 and 5 surface codes. On distances up to 11, the decoder maintains its advantage on simulated data with realistic noise including cross-talk, leakage, and analog readout signals, and sustains its accuracy far beyond the 25 cycles it was trained on. Our work illustrates the ability of machine learning to go beyond human-designed algorithms by learning from data directly, highlighting machine learning as a strong contender for decoding in quantum computers.
Molecular representation learning with language models and domain-relevant auxiliary tasks
Nov 26, 2020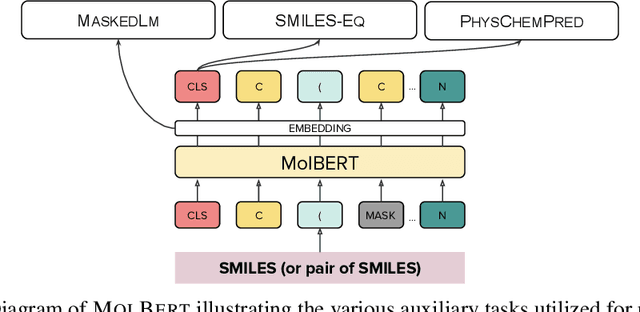
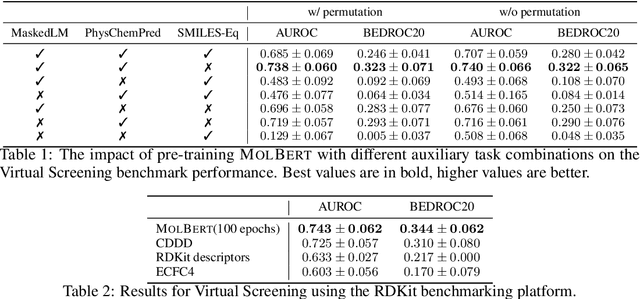
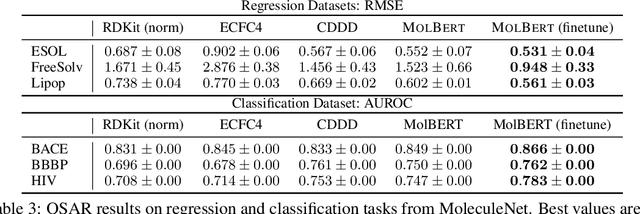
Abstract:We apply a Transformer architecture, specifically BERT, to learn flexible and high quality molecular representations for drug discovery problems. We study the impact of using different combinations of self-supervised tasks for pre-training, and present our results for the established Virtual Screening and QSAR benchmarks. We show that: i) The selection of appropriate self-supervised task(s) for pre-training has a significant impact on performance in subsequent downstream tasks such as Virtual Screening. ii) Using auxiliary tasks with more domain relevance for Chemistry, such as learning to predict calculated molecular properties, increases the fidelity of our learnt representations. iii) Finally, we show that molecular representations learnt by our model `MolBert' improve upon the current state of the art on the benchmark datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge