Electrocardiography Ecg
Papers and Code
Mamba-based Deep Learning Approaches for Sleep Staging on a Wireless Multimodal Wearable System without Electroencephalography
Dec 20, 2024



Study Objectives: We investigate using Mamba-based deep learning approaches for sleep staging on signals from ANNE One (Sibel Health, Evanston, IL), a minimally intrusive dual-sensor wireless wearable system measuring chest electrocardiography (ECG), triaxial accelerometry, and temperature, as well as finger photoplethysmography (PPG) and temperature. Methods: We obtained wearable sensor recordings from 360 adults undergoing concurrent clinical polysomnography (PSG) at a tertiary care sleep lab. PSG recordings were scored according to AASM criteria. PSG and wearable sensor data were automatically aligned using their ECG channels with manual confirmation by visual inspection. We trained Mamba-based models with both convolutional-recurrent neural network (CRNN) and the recurrent neural network (RNN) architectures on these recordings. Ensembling of model variants with similar architectures was performed. Results: Our best approach, after ensembling, attains a 3-class (wake, NREM, REM) balanced accuracy of 83.50%, F1 score of 84.16%, Cohen's $\kappa$ of 72.68%, and a MCC score of 72.84%; a 4-class (wake, N1/N2, N3, REM) balanced accuracy of 74.64%, F1 score of 74.56%, Cohen's $\kappa$ of 61.63%, and MCC score of 62.04%; a 5-class (wake, N1, N2, N3, REM) balanced accuracy of 64.30%, F1 score of 66.97%, Cohen's $\kappa$ of 53.23%, MCC score of 54.38%. Conclusions: Deep learning models can infer major sleep stages from a wearable system without electroencephalography (EEG) and can be successfully applied to data from adults attending a tertiary care sleep clinic.
Bilateral Signal Warping for Left Ventricular Hypertrophy Diagnosis
Nov 13, 2024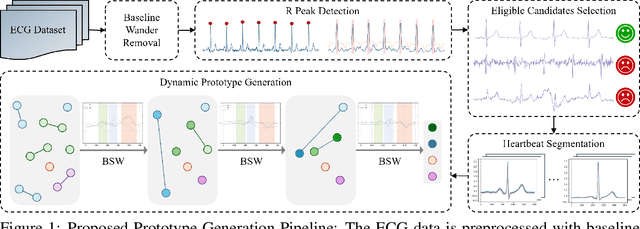
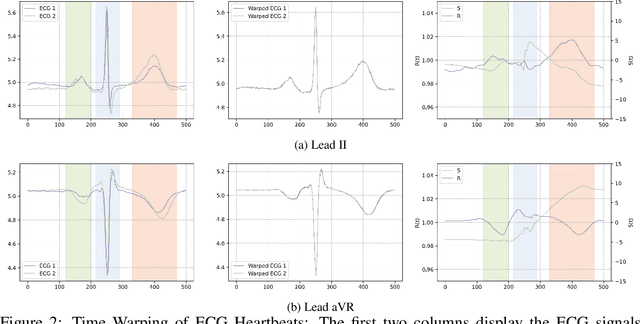
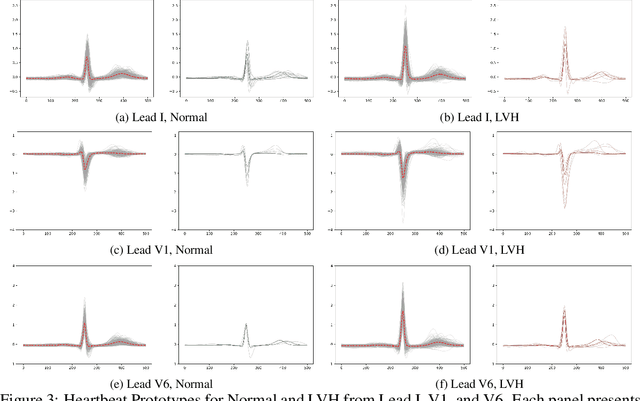
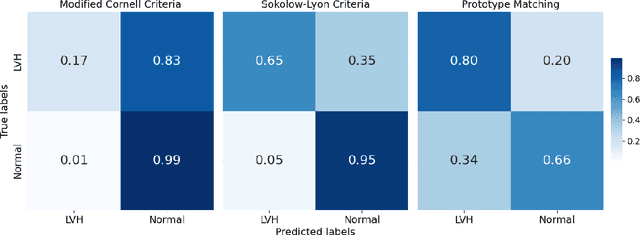
Left Ventricular Hypertrophy (LVH) is a major cardiovascular risk factor, linked to heart failure, arrhythmia, and sudden cardiac death, often resulting from chronic stress like hypertension. Electrocardiography (ECG), while varying in sensitivity, is widely accessible and cost-effective for detecting LVH-related morphological changes. This work introduces a bilateral signal warping (BSW) approach to improve ECG-based LVH diagnosis. Our method creates a library of heartbeat prototypes from patients with consistent ECG patterns. After preprocessing to eliminate baseline wander and detect R peaks, we apply BSW to cluster heartbeats, generating prototypes for both normal and LVH classes. We compare each new record to these references to support diagnosis. Experimental results show promising potential for practical application in clinical settings.
Large-scale cross-modality pretrained model enhances cardiovascular state estimation and cardiomyopathy detection from electrocardiograms: An AI system development and multi-center validation study
Nov 19, 2024



Cardiovascular diseases (CVDs) present significant challenges for early and accurate diagnosis. While cardiac magnetic resonance imaging (CMR) is the gold standard for assessing cardiac function and diagnosing CVDs, its high cost and technical complexity limit accessibility. In contrast, electrocardiography (ECG) offers promise for large-scale early screening. This study introduces CardiacNets, an innovative model that enhances ECG analysis by leveraging the diagnostic strengths of CMR through cross-modal contrastive learning and generative pretraining. CardiacNets serves two primary functions: (1) it evaluates detailed cardiac function indicators and screens for potential CVDs, including coronary artery disease, cardiomyopathy, pericarditis, heart failure and pulmonary hypertension, using ECG input; and (2) it enhances interpretability by generating high-quality CMR images from ECG data. We train and validate the proposed CardiacNets on two large-scale public datasets (the UK Biobank with 41,519 individuals and the MIMIC-IV-ECG comprising 501,172 samples) as well as three private datasets (FAHZU with 410 individuals, SAHZU with 464 individuals, and QPH with 338 individuals), and the findings demonstrate that CardiacNets consistently outperforms traditional ECG-only models, substantially improving screening accuracy. Furthermore, the generated CMR images provide valuable diagnostic support for physicians of all experience levels. This proof-of-concept study highlights how ECG can facilitate cross-modal insights into cardiac function assessment, paving the way for enhanced CVD screening and diagnosis at a population level.
Electromechanical Dynamics of the Heart: A Study of Cardiac Hysteresis During Physical Stress Test
Oct 25, 2024

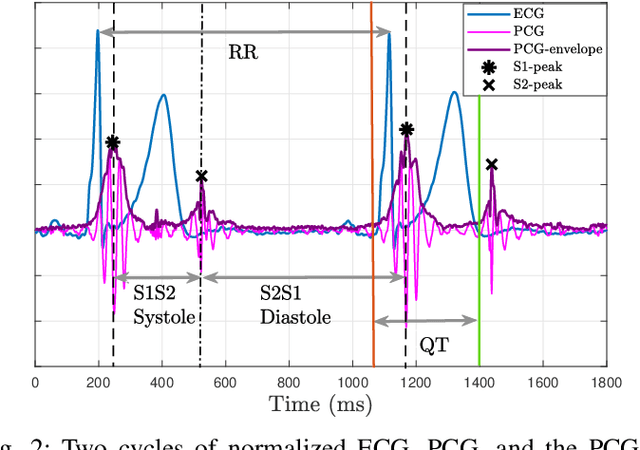

Cardiovascular diseases are best diagnosed using multiple modalities that assess both the heart's electrical and mechanical functions. While effective, imaging techniques like echocardiography and nuclear imaging are costly and not widely accessible. More affordable technologies, such as simultaneous electrocardiography (ECG) and phonocardiography (PCG), may provide valuable insights into electromechanical coupling and could be useful for prescreening in low-resource settings. Using physical stress test data from the EPHNOGRAM ECG-PCG dataset, collected from 23 healthy male subjects (age: 25.4+/-1.9 yrs), we investigated electromechanical intervals (RR, QT, systolic, and diastolic) and their interactions during exercise, along with hysteresis between cardiac electrical activity and mechanical responses. Time delay analysis revealed distinct temporal relationships between QT, systolic, and diastolic intervals, with RR as the primary driver. The diastolic interval showed near-synchrony with RR, while QT responded to RR interval changes with an average delay of 10.5s, and the systolic interval responded more slowly, with an average delay of 28.3s. We examined QT-RR, systolic-RR, and diastolic-RR hysteresis, finding narrower loops for diastolic RR and wider loops for systolic RR. Significant correlations (average:0.75) were found between heart rate changes and hysteresis loop areas, suggesting the equivalent circular area diameter as a promising biomarker for cardiac function under exercise stress. Deep learning models, including Long Short-Term Memory and Convolutional Neural Networks, estimated the QT, systolic, and diastolic intervals from RR data, confirming the nonlinear relationship between RR and other intervals. Findings highlight a significant cardiac memory effect, linking ECG and PCG morphology and timing to heart rate history.
PiEEG-16 to Measure 16 EEG Channels with Raspberry Pi for Brain-Computer Interfaces and EEG devices
Sep 08, 2024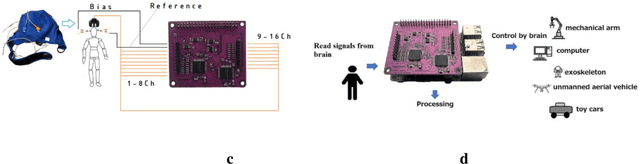
This article introduces a cost-effective gateway into the fascinating world of neuroscience: the PIEEG-16, a versatile shield for RaspberryPi designed to measure 16 channels of various biosignals, including EEG (electroencephalography), EMG (electromyography), and ECG (electrocardiography) without any data transfer over the network (Wi-Fi, Bluetooth) and processing and feature ectraction directly on the Raspberry in real-time. This innovative tool opens up new possibilities for neuroscience research and brain-computer interface experiments. By combining the power of RaspberryPi with specialized biosignal measurement capabilities, the PIEEG-16 represents a significant step forward in democratizing neuroscience research and exploration.
Zodiac: A Cardiologist-Level LLM Framework for Multi-Agent Diagnostics
Oct 02, 2024Large language models (LLMs) have demonstrated remarkable progress in healthcare. However, a significant gap remains regarding LLMs' professionalism in domain-specific clinical practices, limiting their application in real-world diagnostics. In this work, we introduce ZODIAC, an LLM-powered framework with cardiologist-level professionalism designed to engage LLMs in cardiological diagnostics. ZODIAC assists cardiologists by extracting clinically relevant characteristics from patient data, detecting significant arrhythmias, and generating preliminary reports for the review and refinement by cardiologists. To achieve cardiologist-level professionalism, ZODIAC is built on a multi-agent collaboration framework, enabling the processing of patient data across multiple modalities. Each LLM agent is fine-tuned using real-world patient data adjudicated by cardiologists, reinforcing the model's professionalism. ZODIAC undergoes rigorous clinical validation with independent cardiologists, evaluated across eight metrics that measure clinical effectiveness and address security concerns. Results show that ZODIAC outperforms industry-leading models, including OpenAI's GPT-4o, Meta's Llama-3.1-405B, and Google's Gemini-pro, as well as medical-specialist LLMs like Microsoft's BioGPT. ZODIAC demonstrates the transformative potential of specialized LLMs in healthcare by delivering domain-specific solutions that meet the stringent demands of medical practice. Notably, ZODIAC has been successfully integrated into electrocardiography (ECG) devices, exemplifying the growing trend of embedding LLMs into Software-as-Medical-Device (SaMD).
Flexible framework for generating synthetic electrocardiograms and photoplethysmograms
Aug 29, 2024



By generating synthetic biosignals, the quantity and variety of health data can be increased. This is especially useful when training machine learning models by enabling data augmentation and introduction of more physiologically plausible variation to the data. For these purposes, we have developed a synthetic biosignal model for two signal modalities, electrocardiography (ECG) and photoplethysmography (PPG). The model produces realistic signals that account for physiological effects such as breathing modulation and changes in heart rate due to physical stress. Arrhythmic signals can be generated with beat intervals extracted from real measurements. The model also includes a flexible approach to adding different kinds of noise and signal artifacts. The noise is generated from power spectral densities extracted from both measured noisy signals and modeled power spectra. Importantly, the model also automatically produces labels for noise, segmentation (e.g. P and T waves, QRS complex, for electrocardiograms), and artifacts. We assessed how this comprehensive model can be used in practice to improve the performance of models trained on ECG or PPG data. For example, we trained an LSTM to detect ECG R-peaks using both real ECG signals from the MIT-BIH arrythmia set and our new generator. The F1 score of the model was 0.83 using real data, in comparison to 0.98 using our generator. In addition, the model can be used for example in signal segmentation, quality detection and bench-marking detection algorithms. The model code has been released in \url{https://github.com/UTU-Health-Research/framework_for_synthetic_biosignals}
Workload Assessment of Human-Machine Interface: A Simulator Study with Psychophysiological Measures
Jun 13, 2024Human-machine Interface (HMI) is critical for safety during automated driving, as it serves as the only media between the automated system and human users. To enable a transparent HMI, we first need to know how to evaluate it. However, most of the assessment methods used for HMI designs are subjective and thus not efficient. To bridge the gap, an objective and standardized HMI assessment method is needed, and the first step is to find an objective method for workload measurement for this context. In this study, two psychophysiological measures, electrocardiography (ECG) and electrodermal activity (EDA), were evaluated for their effectiveness in finding differences in mental workload among different HMI designs in a simulator study. Three HMI designs were developed and used. Results showed that both workload measures were able to identify significant differences in objective mental workload when interacting with in-vehicle HMIs. As a first step toward a standardized assessment method, the results could be used as a firm ground for future studies. Marie Sk{\l}odowska-Curie Actions; Innovative Training Network (ITN); SHAPE-IT; Grant number 860410; Publication date: [29 Sep 2023]; DOI: [10.54941/ahfe1004172]
Computation-Efficient Semi-Supervised Learning for ECG-based Cardiovascular Diseases Detection
Jun 20, 2024Label scarcity problem is the main challenge that hinders the wide application of deep learning systems in automatic cardiovascular diseases (CVDs) detection using electrocardiography (ECG). Tuning pre-trained models alleviates this problem by transferring knowledge learned from large datasets to downstream small datasets. However, bottlenecks in computational efficiency and CVDs detection performance limit its clinical applications. It is difficult to improve the detection performance without significantly sacrificing model computational efficiency. Here, we propose a computation-efficient semi-supervised learning paradigm (FastECG) for robust and computation-efficient CVDs detection using ECG. It enables a robust adaptation of pre-trained models on downstream datasets with limited supervision and high computational efficiency. First, a random-deactivation technique is developed to achieve robust and fast low-rank adaptation of pre-trained weights. Subsequently, we propose a one-shot rank allocation module to determine the optimal ranks for the update matrices of the pre-trained weights. Finally, a lightweight semi-supervised learning pipeline is introduced to enhance model performance by leveraging labeled and unlabeled data with high computational efficiency. Extensive experiments on four downstream ECG datasets demonstrate that FastECG not only outperforms the state-of-the-art methods in multi-label CVDs detection but also consumes fewer GPU footprints, training time, and parameter storage space. As such, this paradigm provides an effective solution for achieving high computational efficiency and robust detection performance in the clinical applications of pre-trained models under limited supervision.
Uncertainty-Aware PPG-2-ECG for Enhanced Cardiovascular Diagnosis using Diffusion Models
May 19, 2024Analyzing the cardiovascular system condition via Electrocardiography (ECG) is a common and highly effective approach, and it has been practiced and perfected over many decades. ECG sensing is non-invasive and relatively easy to acquire, and yet it is still cumbersome for holter monitoring tests that may span over hours and even days. A possible alternative in this context is Photoplethysmography (PPG): An optically-based signal that measures blood volume fluctuations, as typically sensed by conventional ``wearable devices''. While PPG presents clear advantages in acquisition, convenience, and cost-effectiveness, ECG provides more comprehensive information, allowing for a more precise detection of heart conditions. This implies that a conversion from PPG to ECG, as recently discussed in the literature, inherently involves an unavoidable level of uncertainty. In this paper we introduce a novel methodology for addressing the PPG-2-ECG conversion, and offer an enhanced classification of cardiovascular conditions using the given PPG, all while taking into account the uncertainties arising from the conversion process. We provide a mathematical justification for our proposed computational approach, and present empirical studies demonstrating its superior performance compared to state-of-the-art baseline methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge