Antti Airola
Interaction Concordance Index: Performance Evaluation for Interaction Prediction Methods
Oct 16, 2025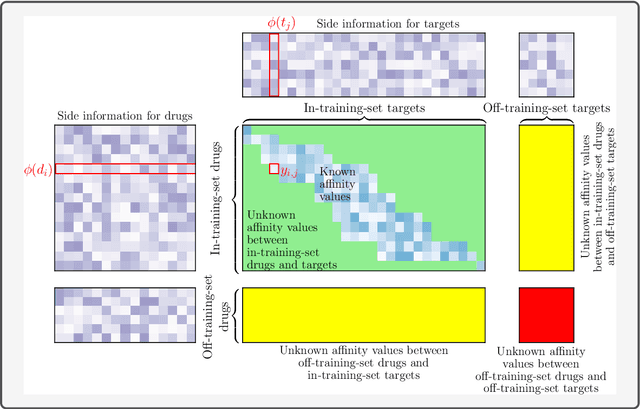

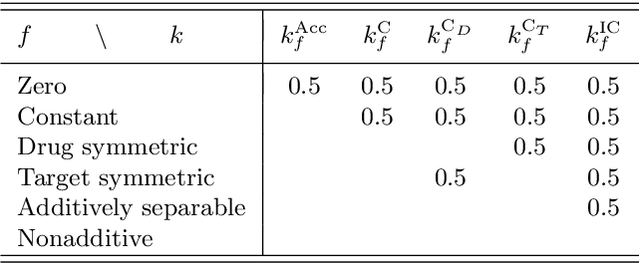
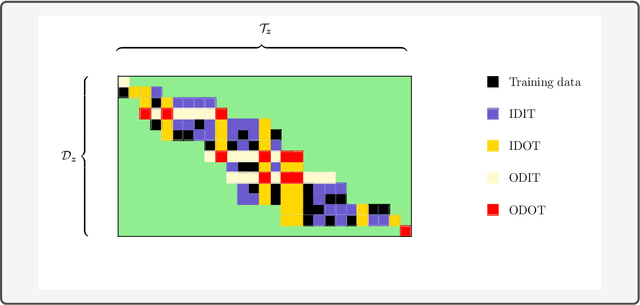
Abstract:Consider two sets of entities and their members' mutual affinity values, say drug-target affinities (DTA). Drugs and targets are said to interact in their effects on DTAs if drug's effect on it depends on the target. Presence of interaction implies that assigning a drug to a target and another drug to another target does not provide the same aggregate DTA as the reversed assignment would provide. Accordingly, correctly capturing interactions enables better decision-making, for example, in allocation of limited numbers of drug doses to their best matching targets. Learning to predict DTAs is popularly done from either solely from known DTAs or together with side information on the entities, such as chemical structures of drugs and targets. In this paper, we introduce interaction directions' prediction performance estimator we call interaction concordance index (IC-index), for both fixed predictors and machine learning algorithms aimed for inferring them. IC-index complements the popularly used DTA prediction performance estimators by evaluating the ratio of correctly predicted directions of interaction effects in data. First, we show the invariance of IC-index on predictors unable to capture interactions. Secondly, we show that learning algorithm's permutation equivariance regarding drug and target identities implies its inability to capture interactions when either drug, target or both are unseen during training. In practical applications, this equivariance is remedied via incorporation of appropriate side information on drugs and targets. We make a comprehensive empirical evaluation over several biomedical interaction data sets with various state-of-the-art machine learning algorithms. The experiments demonstrate how different types of affinity strength prediction methods perform in terms of IC-index complementing existing prediction performance estimators.
A Comprehensive Guide to Differential Privacy: From Theory to User Expectations
Sep 03, 2025Abstract:The increasing availability of personal data has enabled significant advances in fields such as machine learning, healthcare, and cybersecurity. However, this data abundance also raises serious privacy concerns, especially in light of powerful re-identification attacks and growing legal and ethical demands for responsible data use. Differential privacy (DP) has emerged as a principled, mathematically grounded framework for mitigating these risks. This review provides a comprehensive survey of DP, covering its theoretical foundations, practical mechanisms, and real-world applications. It explores key algorithmic tools and domain-specific challenges - particularly in privacy-preserving machine learning and synthetic data generation. The report also highlights usability issues and the need for improved communication and transparency in DP systems. Overall, the goal is to support informed adoption of DP by researchers and practitioners navigating the evolving landscape of data privacy.
Flexible framework for generating synthetic electrocardiograms and photoplethysmograms
Aug 29, 2024



Abstract:By generating synthetic biosignals, the quantity and variety of health data can be increased. This is especially useful when training machine learning models by enabling data augmentation and introduction of more physiologically plausible variation to the data. For these purposes, we have developed a synthetic biosignal model for two signal modalities, electrocardiography (ECG) and photoplethysmography (PPG). The model produces realistic signals that account for physiological effects such as breathing modulation and changes in heart rate due to physical stress. Arrhythmic signals can be generated with beat intervals extracted from real measurements. The model also includes a flexible approach to adding different kinds of noise and signal artifacts. The noise is generated from power spectral densities extracted from both measured noisy signals and modeled power spectra. Importantly, the model also automatically produces labels for noise, segmentation (e.g. P and T waves, QRS complex, for electrocardiograms), and artifacts. We assessed how this comprehensive model can be used in practice to improve the performance of models trained on ECG or PPG data. For example, we trained an LSTM to detect ECG R-peaks using both real ECG signals from the MIT-BIH arrythmia set and our new generator. The F1 score of the model was 0.83 using real data, in comparison to 0.98 using our generator. In addition, the model can be used for example in signal segmentation, quality detection and bench-marking detection algorithms. The model code has been released in \url{https://github.com/UTU-Health-Research/framework_for_synthetic_biosignals}
Empirical investigation of multi-source cross-validation in clinical machine learning
Mar 22, 2024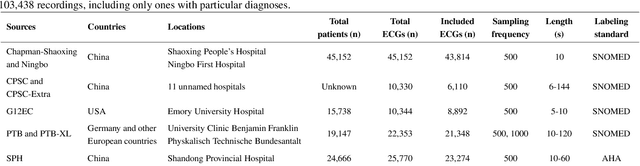
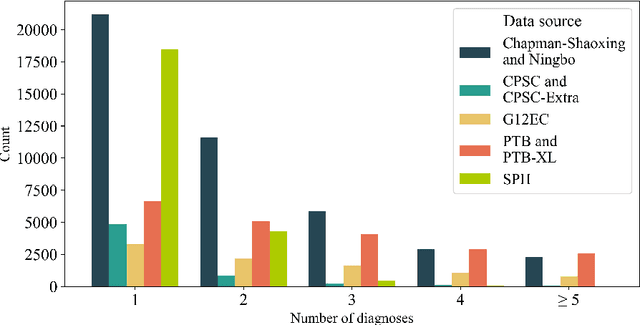
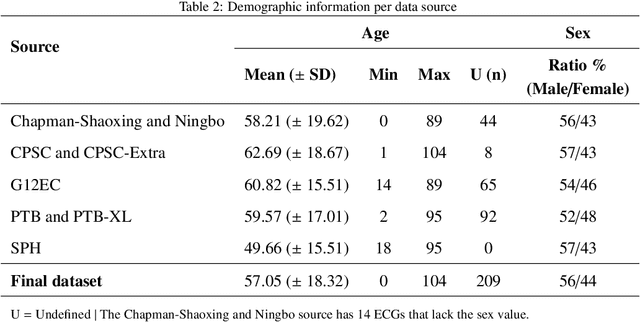
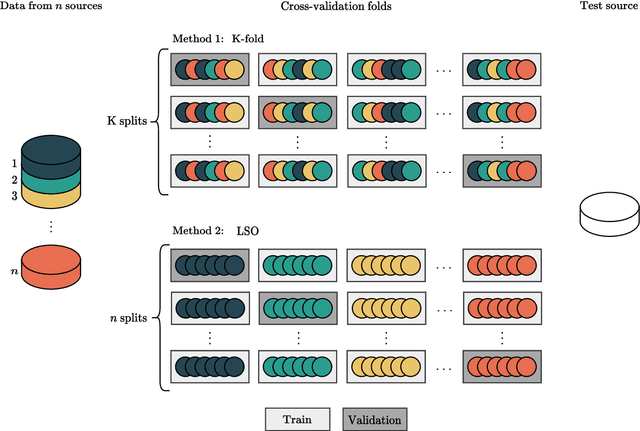
Abstract:Traditionally, machine learning-based clinical prediction models have been trained and evaluated on patient data from a single source, such as a hospital. Cross-validation methods can be used to estimate the accuracy of such models on new patients originating from the same source, by repeated random splitting of the data. However, such estimates tend to be highly overoptimistic when compared to accuracy obtained from deploying models to sources not represented in the dataset, such as a new hospital. The increasing availability of multi-source medical datasets provides new opportunities for obtaining more comprehensive and realistic evaluations of expected accuracy through source-level cross-validation designs. In this study, we present a systematic empirical evaluation of standard K-fold cross-validation and leave-source-out cross-validation methods in a multi-source setting. We consider the task of electrocardiogram based cardiovascular disease classification, combining and harmonizing the openly available PhysioNet CinC Challenge 2021 and the Shandong Provincial Hospital datasets for our study. Our results show that K-fold cross-validation, both on single-source and multi-source data, systemically overestimates prediction performance when the end goal is to generalize to new sources. Leave-source-out cross-validation provides more reliable performance estimates, having close to zero bias though larger variability. The evaluation highlights the dangers of obtaining misleading cross-validation results on medical data and demonstrates how these issues can be mitigated when having access to multi-source data.
Does Differentially Private Synthetic Data Lead to Synthetic Discoveries?
Mar 20, 2024Abstract:Background: Synthetic data has been proposed as a solution for sharing anonymized versions of sensitive biomedical datasets. Ideally, synthetic data should preserve the structure and statistical properties of the original data, while protecting the privacy of the individual subjects. Differential privacy (DP) is currently considered the gold standard approach for balancing this trade-off. Objectives: The aim of this study is to evaluate the Mann-Whitney U test on DP-synthetic biomedical data in terms of Type I and Type II errors, in order to establish whether statistical hypothesis testing performed on privacy preserving synthetic data is likely to lead to loss of test's validity or decreased power. Methods: We evaluate the Mann-Whitney U test on DP-synthetic data generated from real-world data, including a prostate cancer dataset (n=500) and a cardiovascular dataset (n=70 000), as well as on data drawn from two Gaussian distributions. Five different DP-synthetic data generation methods are evaluated, including two basic DP histogram release methods and MWEM, Private-PGM, and DP GAN algorithms. Conclusion: Most of the tested DP-synthetic data generation methods showed inflated Type I error, especially at privacy budget levels of $\epsilon\leq 1$. This result calls for caution when releasing and analyzing DP-synthetic data: low p-values may be obtained in statistical tests simply as a byproduct of the noise added to protect privacy. A DP smoothed histogram-based synthetic data generation method was shown to produce valid Type I error for all privacy levels tested but required a large original dataset size and a modest privacy budget ($\epsilon\geq 5$) in order to have reasonable Type II error levels.
Training neural networks with synthetic electrocardiograms
Nov 11, 2021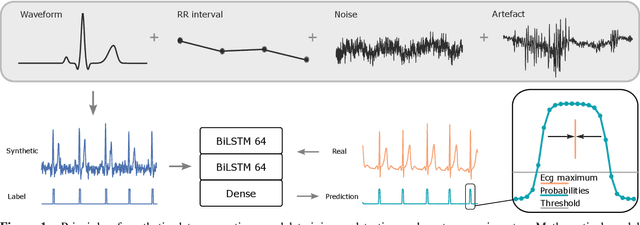
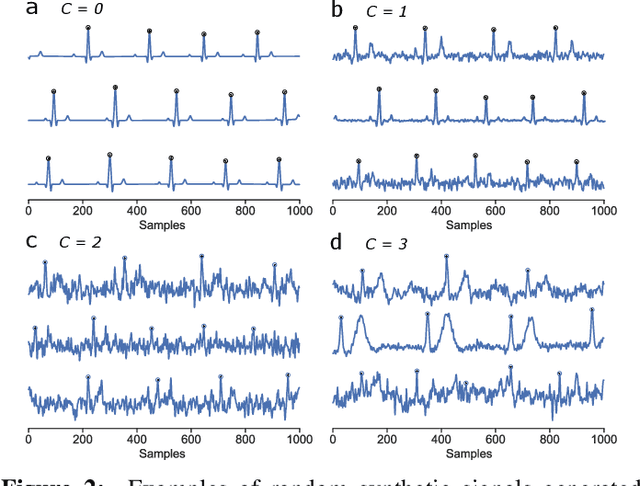
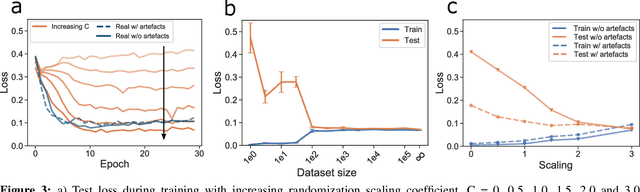
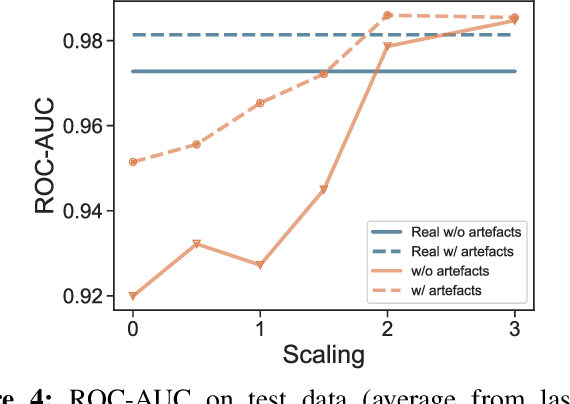
Abstract:We present a method for training neural networks with synthetic electrocardiograms that mimic signals produced by a wearable single lead electrocardiogram monitor. We use domain randomization where the synthetic signal properties such as the waveform shape, RR-intervals and noise are varied for every training example. Models trained with synthetic data are compared to their counterparts trained with real data. Detection of r-waves in electrocardiograms recorded during different physical activities and in atrial fibrillation is used to compare the models. By allowing the randomization to increase beyond what is typically observed in the real-world data the performance is on par or superseding the performance of networks trained with real data. Experiments show robust performance with different seeds and training examples on different test sets without any test set specific tuning. The method makes possible to train neural networks using practically free-to-collect data with accurate labels without the need for manual annotations and it opens up the possibility of extending the use of synthetic data on cardiac disease classification when disease specific a priori information is used in the electrocardiogram generation. Additionally the distribution of data can be controlled eliminating class imbalances that are typically observed in health related data and additionally the generated data is inherently private.
A Link between Coding Theory and Cross-Validation with Applications
Mar 22, 2021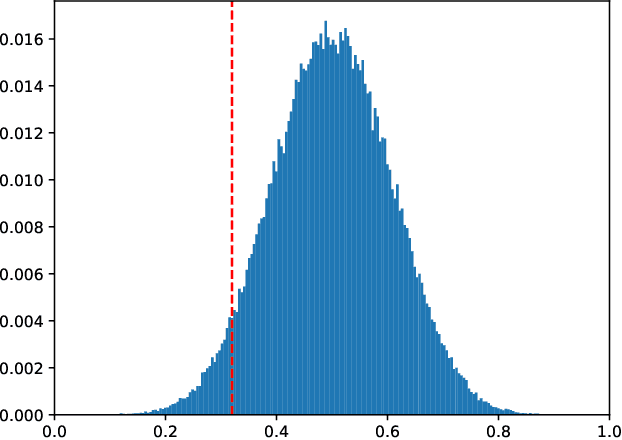
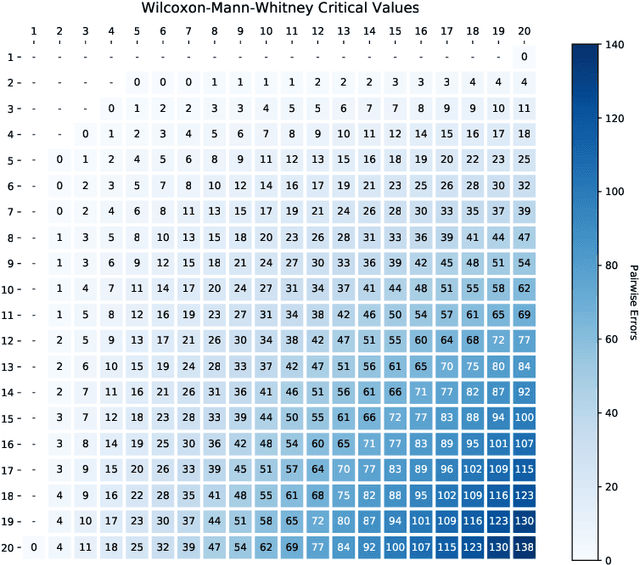
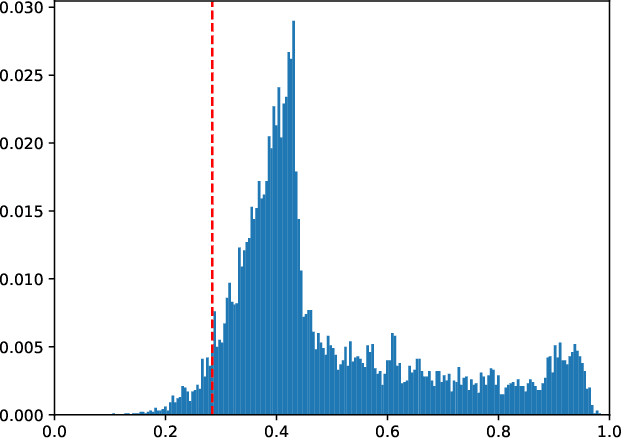
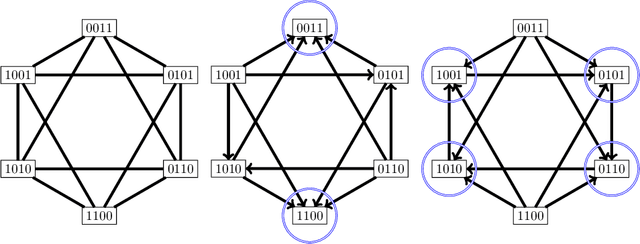
Abstract:We study the combinatorics of cross-validation based AUC estimation under the null hypothesis that the binary class labels are exchangeable, that is, the data are randomly assigned into two classes given a fixed class proportion. In particular, we study how the estimators based on leave-pair-out cross-validation (LPOCV), in which every possible pair of data with different class labels is held out from the training set at a time, behave under the null without any prior assumptions of the learning algorithm or the data. It is shown that the maximal number of different fixed proportion label assignments on a sample of data, for which a learning algorithm can achieve zero LPOCV error, is the maximal size of a constant weight error correcting code, whose length is the sample size, weight is the number of data labeled with one, and the Hamming distance between code words is four. We then introduce the concept of a light constant weight code and show similar results for nonzero LPOCV errors. We also prove both upper and lower bounds on the maximal sizes of the light constant weight codes that are similar to the classical results for contant weight codes. These results pave the way towards the design of new LPOCV based statistical tests for the learning algorithms ability of distinguishing two classes from each other that are analogous to the classical Wilcoxon-Mann-Whitney U test for fixed functions. Behavior of some representative examples of learning algorithms and data are simulated in an experimental case study.
Generalized vec trick for fast learning of pairwise kernel models
Sep 02, 2020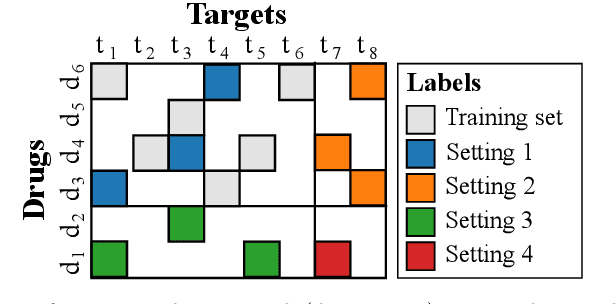
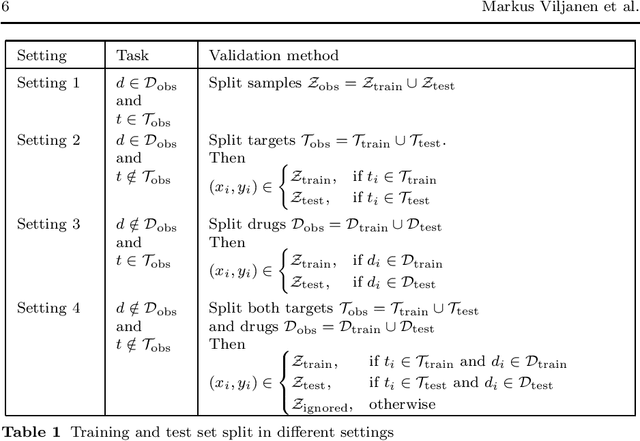

Abstract:Pairwise learning corresponds to the supervised learning setting where the goal is to make predictions for pairs of objects. Prominent applications include predicting drug-target or protein-protein interactions, or customer-product preferences. Several kernel functions have been proposed for incorporating prior knowledge about the relationship between the objects, when training kernel based learning methods. However, the number of training pairs n is often very large, making O(n^2) cost of constructing the pairwise kernel matrix infeasible. If each training pair x= (d,t) consists of drug d and target t, let m and q denote the number of unique drugs and targets appearing in the training pairs. In many real-world applications m,q << n, which can be used to develop computational shortcuts. Recently, a O(nm+nq) time algorithm we refer to as the generalized vec trick was introduced for training kernel methods with the Kronecker kernel. In this work, we show that a large class of pairwise kernels can be expressed as a sum of product matrices, which generalizes the result to the most commonly used pairwise kernels. This includes symmetric and anti-symmetric, metric-learning, Cartesian, ranking, as well as linear, polynomial and Gaussian kernels. In the experiments, we demonstrate how the introduced approach allows scaling pairwise kernels to much larger data sets than previously feasible, and compare the kernels on a number of biological interaction prediction tasks.
A Comparative Study of Pairwise Learning Methods based on Kernel Ridge Regression
Mar 05, 2018Abstract:Many machine learning problems can be formulated as predicting labels for a pair of objects. Problems of that kind are often referred to as pairwise learning, dyadic prediction or network inference problems. During the last decade kernel methods have played a dominant role in pairwise learning. They still obtain a state-of-the-art predictive performance, but a theoretical analysis of their behavior has been underexplored in the machine learning literature. In this work we review and unify existing kernel-based algorithms that are commonly used in different pairwise learning settings, ranging from matrix filtering to zero-shot learning. To this end, we focus on closed-form efficient instantiations of Kronecker kernel ridge regression. We show that independent task kernel ridge regression, two-step kernel ridge regression and a linear matrix filter arise naturally as a special case of Kronecker kernel ridge regression, implying that all these methods implicitly minimize a squared loss. In addition, we analyze universality, consistency and spectral filtering properties. Our theoretical results provide valuable insights in assessing the advantages and limitations of existing pairwise learning methods.
Tournament Leave-pair-out Cross-validation for Receiver Operating Characteristic (ROC) Analysis
Jan 29, 2018
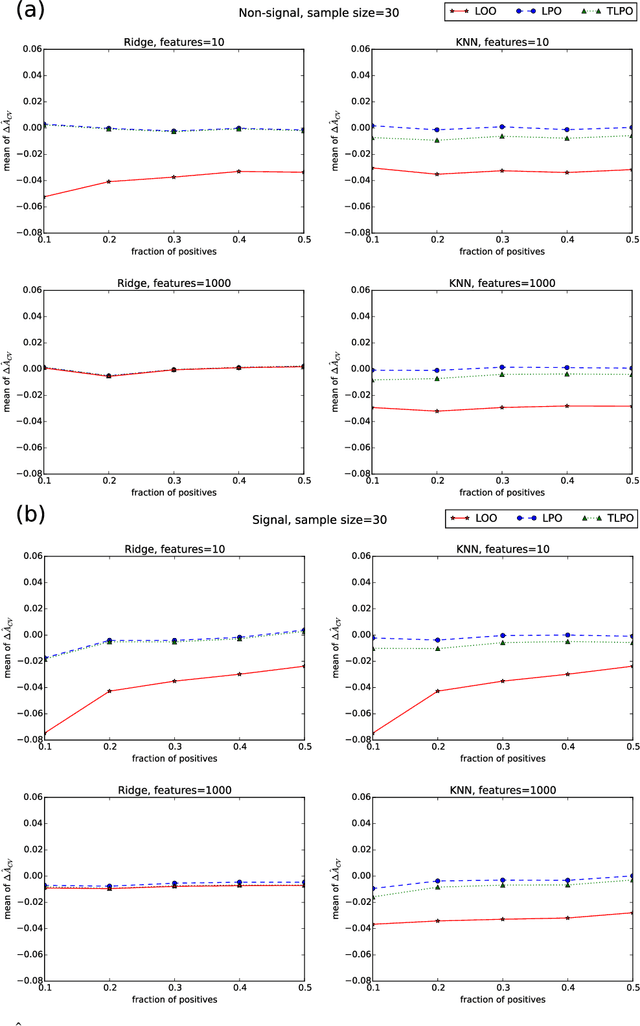
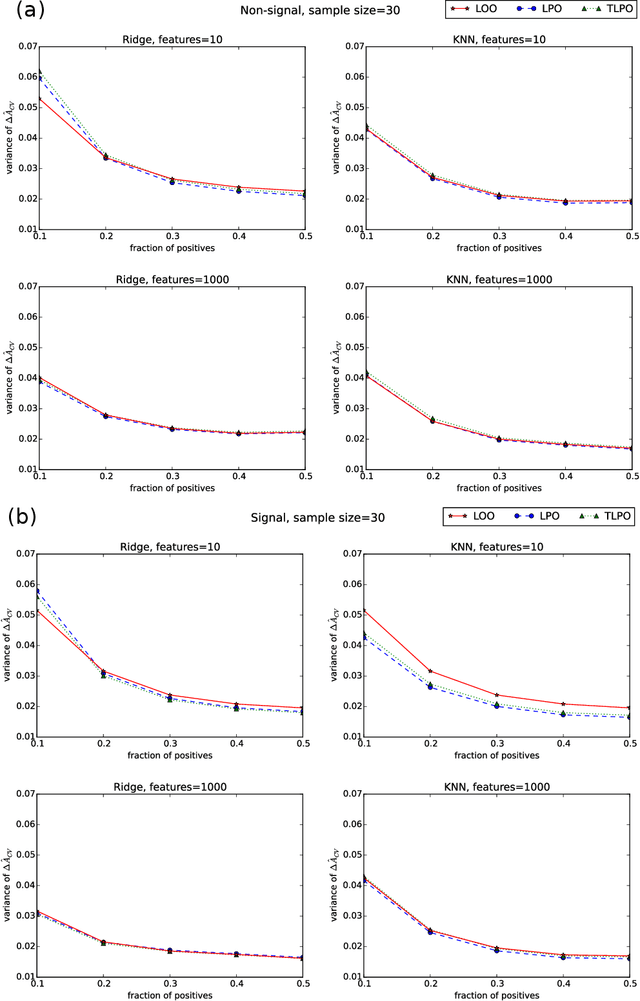

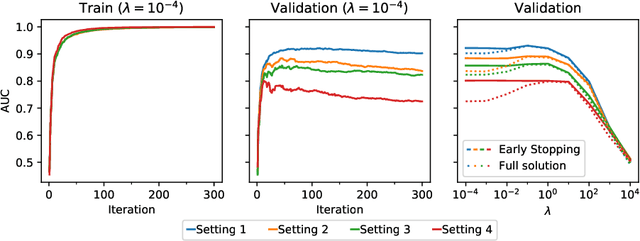
Abstract:Receiver operating characteristic (ROC) analysis is widely used for evaluating diagnostic systems. Recent studies have shown that estimating an area under ROC curve (AUC) with standard cross-validation methods suffers from a large bias. The leave-pair-out (LPO) cross-validation has been shown to correct this bias. However, while LPO produces an almost unbiased estimate of AUC, it does not provide a ranking of the data needed for plotting and analyzing the ROC curve. In this study, we propose a new method called tournament leave-pair-out (TLPO) cross-validation. This method extends LPO by creating a tournament from pair comparisons to produce a ranking for the data. TLPO preserves the advantage of LPO for estimating AUC, while it also allows performing ROC analysis. We have shown using both synthetic and real world data that TLPO is as reliable as LPO for AUC estimation and confirmed the bias in leave-one-out cross-validation on low-dimensional data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge