Ivan Jambor
Cross-Vendor Reproducibility of Radiomics-based Machine Learning Models for Computer-aided Diagnosis
Jul 25, 2024



Abstract:Background: The reproducibility of machine-learning models in prostate cancer detection across different MRI vendors remains a significant challenge. Methods: This study investigates Support Vector Machines (SVM) and Random Forest (RF) models trained on radiomic features extracted from T2-weighted MRI images using Pyradiomics and MRCradiomics libraries. Feature selection was performed using the maximum relevance minimum redundancy (MRMR) technique. We aimed to enhance clinical decision support through multimodal learning and feature fusion. Results: Our SVM model, utilizing combined features from Pyradiomics and MRCradiomics, achieved an AUC of 0.74 on the Multi-Improd dataset (Siemens scanner) but decreased to 0.60 on the Philips test set. The RF model showed similar trends, with notable robustness for models using Pyradiomics features alone (AUC of 0.78 on Philips). Conclusions: These findings demonstrate the potential of multimodal feature integration to improve the robustness and generalizability of machine-learning models for clinical decision support in prostate cancer detection. This study marks a significant step towards developing reliable AI-driven diagnostic tools that maintain efficacy across various imaging platforms.
A Link between Coding Theory and Cross-Validation with Applications
Mar 22, 2021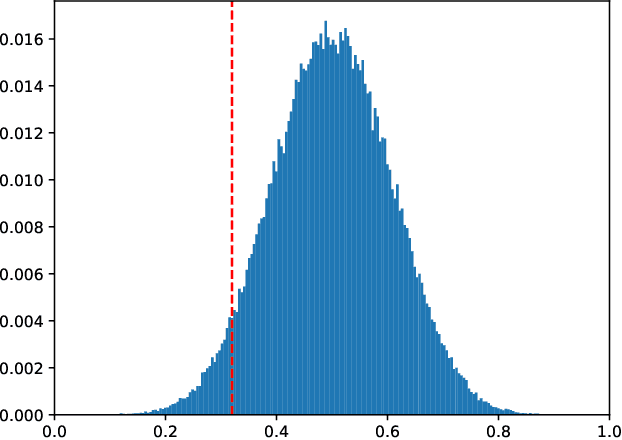
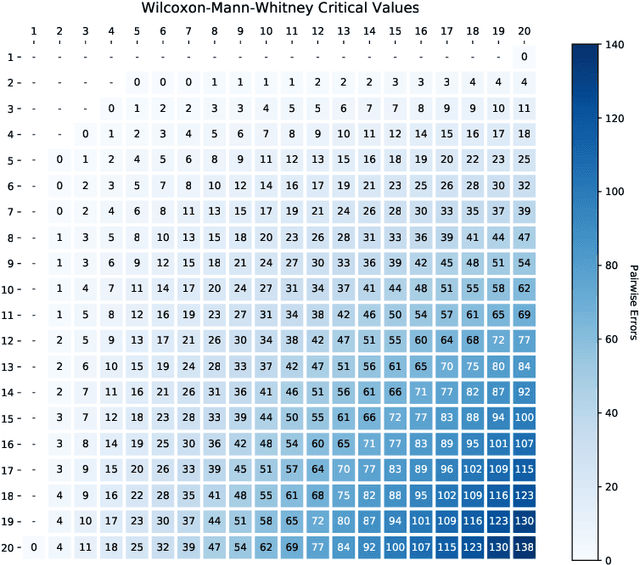
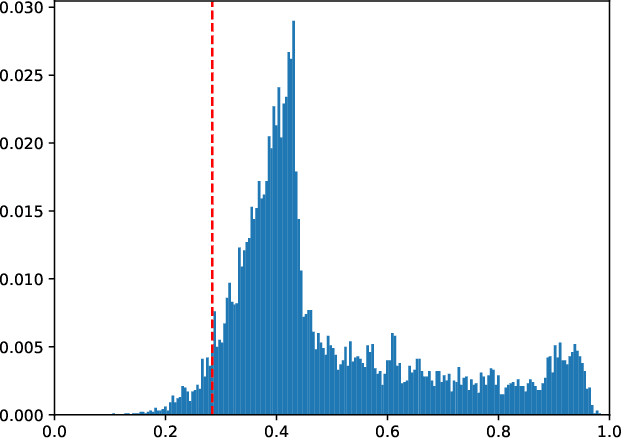
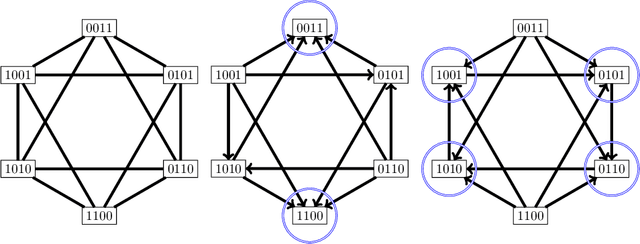
Abstract:We study the combinatorics of cross-validation based AUC estimation under the null hypothesis that the binary class labels are exchangeable, that is, the data are randomly assigned into two classes given a fixed class proportion. In particular, we study how the estimators based on leave-pair-out cross-validation (LPOCV), in which every possible pair of data with different class labels is held out from the training set at a time, behave under the null without any prior assumptions of the learning algorithm or the data. It is shown that the maximal number of different fixed proportion label assignments on a sample of data, for which a learning algorithm can achieve zero LPOCV error, is the maximal size of a constant weight error correcting code, whose length is the sample size, weight is the number of data labeled with one, and the Hamming distance between code words is four. We then introduce the concept of a light constant weight code and show similar results for nonzero LPOCV errors. We also prove both upper and lower bounds on the maximal sizes of the light constant weight codes that are similar to the classical results for contant weight codes. These results pave the way towards the design of new LPOCV based statistical tests for the learning algorithms ability of distinguishing two classes from each other that are analogous to the classical Wilcoxon-Mann-Whitney U test for fixed functions. Behavior of some representative examples of learning algorithms and data are simulated in an experimental case study.
Accurate Prostate Cancer Detection and Segmentation on Biparametric MRI using Non-local Mask R-CNN with Histopathological Ground Truth
Oct 28, 2020



Abstract:Purpose: We aimed to develop deep machine learning (DL) models to improve the detection and segmentation of intraprostatic lesions (IL) on bp-MRI by using whole amount prostatectomy specimen-based delineations. We also aimed to investigate whether transfer learning and self-training would improve results with small amount labelled data. Methods: 158 patients had suspicious lesions delineated on MRI based on bp-MRI, 64 patients had ILs delineated on MRI based on whole mount prostatectomy specimen sections, 40 patients were unlabelled. A non-local Mask R-CNN was proposed to improve the segmentation accuracy. Transfer learning was investigated by fine-tuning a model trained using MRI-based delineations with prostatectomy-based delineations. Two label selection strategies were investigated in self-training. The performance of models was evaluated by 3D detection rate, dice similarity coefficient (DSC), 95 percentile Hausdrauff (95 HD, mm) and true positive ratio (TPR). Results: With prostatectomy-based delineations, the non-local Mask R-CNN with fine-tuning and self-training significantly improved all evaluation metrics. For the model with the highest detection rate and DSC, 80.5% (33/41) of lesions in all Gleason Grade Groups (GGG) were detected with DSC of 0.548[0.165], 95 HD of 5.72[3.17] and TPR of 0.613[0.193]. Among them, 94.7% (18/19) of lesions with GGG > 2 were detected with DSC of 0.604[0.135], 95 HD of 6.26[3.44] and TPR of 0.580[0.190]. Conclusion: DL models can achieve high prostate cancer detection and segmentation accuracy on bp-MRI based on annotations from histologic images. To further improve the performance, more data with annotations of both MRI and whole amount prostatectomy specimens are required.
Tournament Leave-pair-out Cross-validation for Receiver Operating Characteristic (ROC) Analysis
Jan 29, 2018
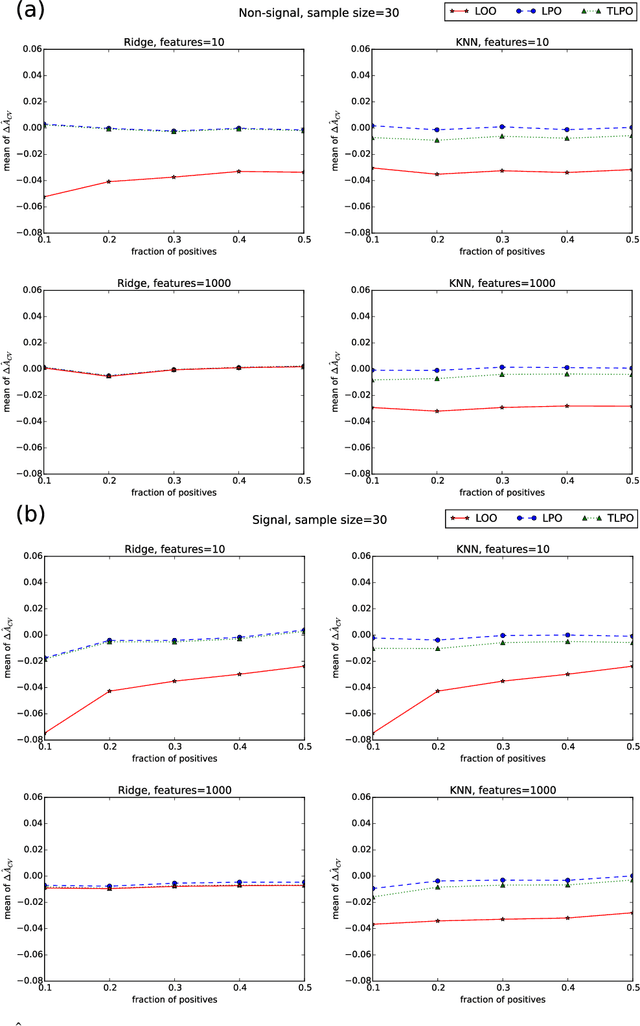
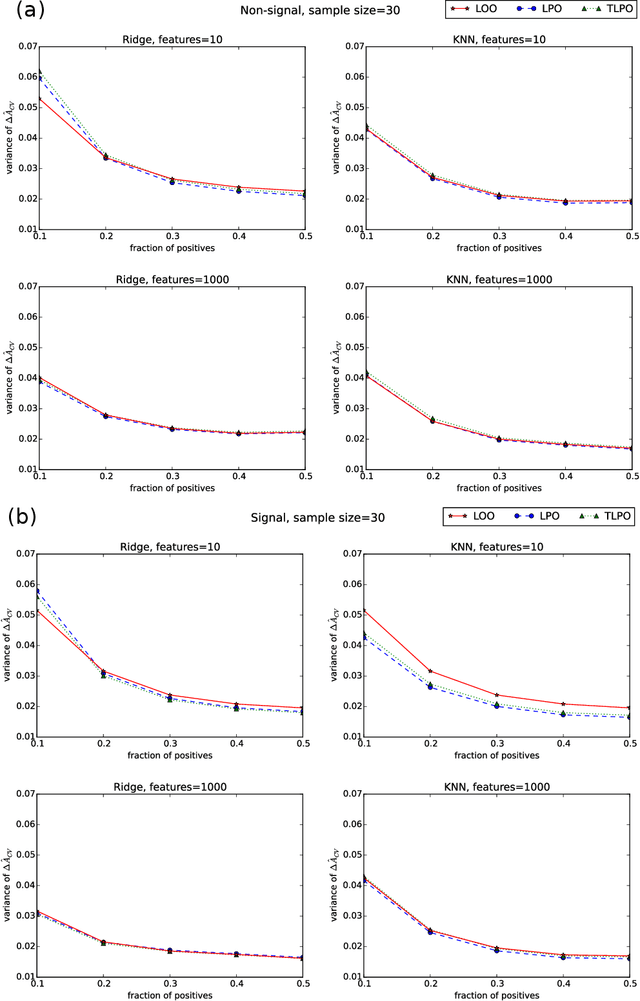

Abstract:Receiver operating characteristic (ROC) analysis is widely used for evaluating diagnostic systems. Recent studies have shown that estimating an area under ROC curve (AUC) with standard cross-validation methods suffers from a large bias. The leave-pair-out (LPO) cross-validation has been shown to correct this bias. However, while LPO produces an almost unbiased estimate of AUC, it does not provide a ranking of the data needed for plotting and analyzing the ROC curve. In this study, we propose a new method called tournament leave-pair-out (TLPO) cross-validation. This method extends LPO by creating a tournament from pair comparisons to produce a ranking for the data. TLPO preserves the advantage of LPO for estimating AUC, while it also allows performing ROC analysis. We have shown using both synthetic and real world data that TLPO is as reliable as LPO for AUC estimation and confirmed the bias in leave-one-out cross-validation on low-dimensional data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge