Ileana Montoya Perez
Does Differentially Private Synthetic Data Lead to Synthetic Discoveries?
Mar 20, 2024Abstract:Background: Synthetic data has been proposed as a solution for sharing anonymized versions of sensitive biomedical datasets. Ideally, synthetic data should preserve the structure and statistical properties of the original data, while protecting the privacy of the individual subjects. Differential privacy (DP) is currently considered the gold standard approach for balancing this trade-off. Objectives: The aim of this study is to evaluate the Mann-Whitney U test on DP-synthetic biomedical data in terms of Type I and Type II errors, in order to establish whether statistical hypothesis testing performed on privacy preserving synthetic data is likely to lead to loss of test's validity or decreased power. Methods: We evaluate the Mann-Whitney U test on DP-synthetic data generated from real-world data, including a prostate cancer dataset (n=500) and a cardiovascular dataset (n=70 000), as well as on data drawn from two Gaussian distributions. Five different DP-synthetic data generation methods are evaluated, including two basic DP histogram release methods and MWEM, Private-PGM, and DP GAN algorithms. Conclusion: Most of the tested DP-synthetic data generation methods showed inflated Type I error, especially at privacy budget levels of $\epsilon\leq 1$. This result calls for caution when releasing and analyzing DP-synthetic data: low p-values may be obtained in statistical tests simply as a byproduct of the noise added to protect privacy. A DP smoothed histogram-based synthetic data generation method was shown to produce valid Type I error for all privacy levels tested but required a large original dataset size and a modest privacy budget ($\epsilon\geq 5$) in order to have reasonable Type II error levels.
Tournament Leave-pair-out Cross-validation for Receiver Operating Characteristic (ROC) Analysis
Jan 29, 2018
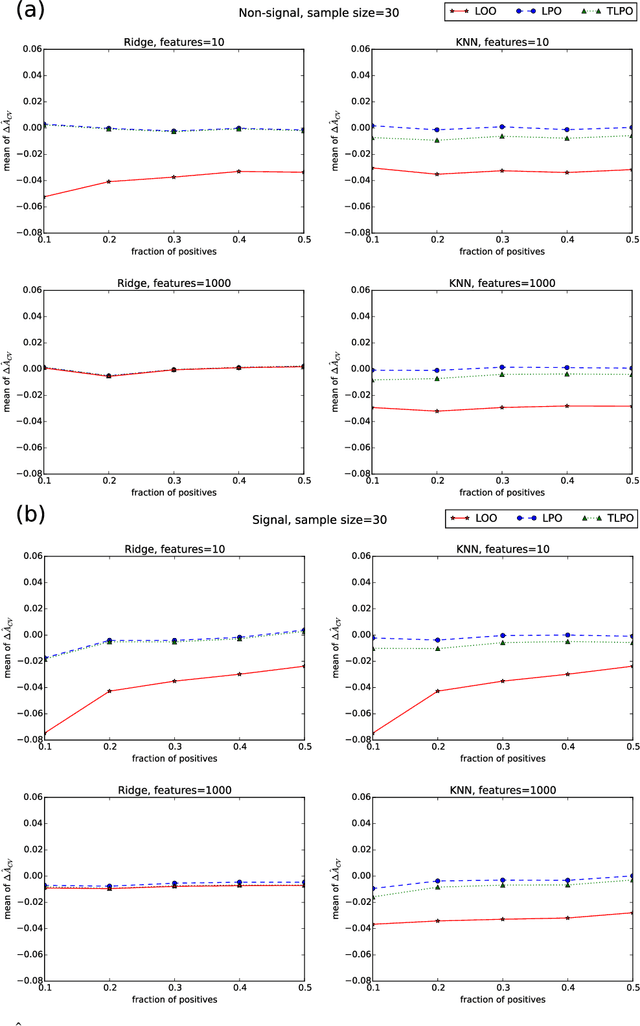
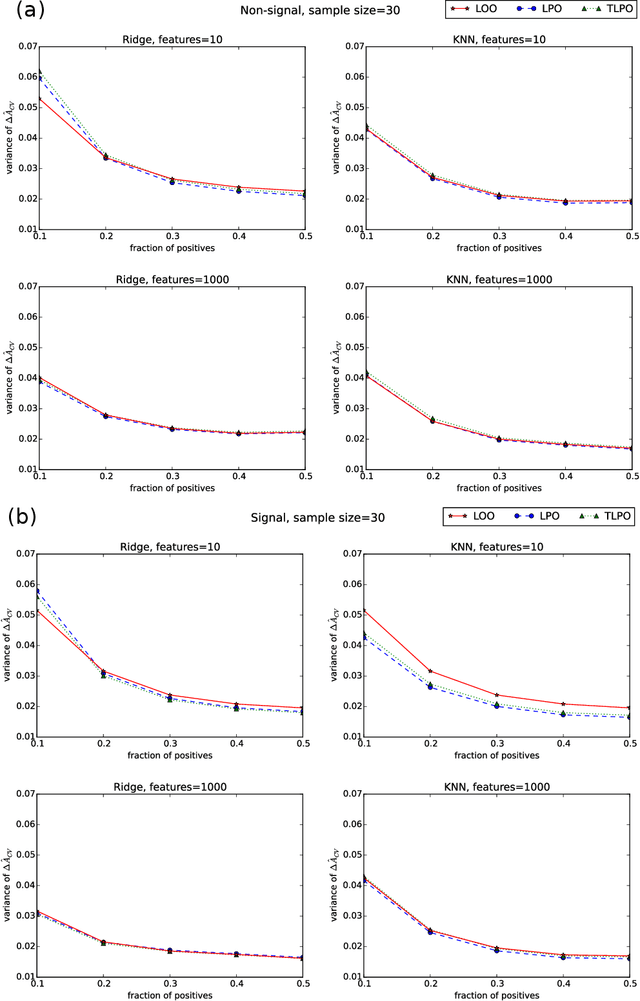

Abstract:Receiver operating characteristic (ROC) analysis is widely used for evaluating diagnostic systems. Recent studies have shown that estimating an area under ROC curve (AUC) with standard cross-validation methods suffers from a large bias. The leave-pair-out (LPO) cross-validation has been shown to correct this bias. However, while LPO produces an almost unbiased estimate of AUC, it does not provide a ranking of the data needed for plotting and analyzing the ROC curve. In this study, we propose a new method called tournament leave-pair-out (TLPO) cross-validation. This method extends LPO by creating a tournament from pair comparisons to produce a ranking for the data. TLPO preserves the advantage of LPO for estimating AUC, while it also allows performing ROC analysis. We have shown using both synthetic and real world data that TLPO is as reliable as LPO for AUC estimation and confirmed the bias in leave-one-out cross-validation on low-dimensional data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge