Empirical investigation of multi-source cross-validation in clinical machine learning
Paper and Code
Mar 22, 2024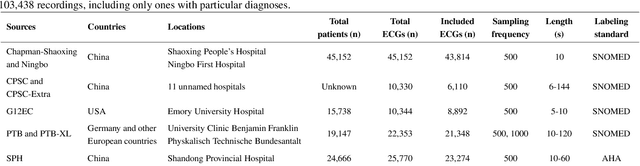
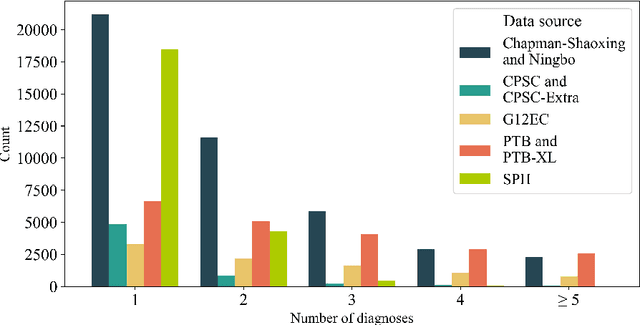
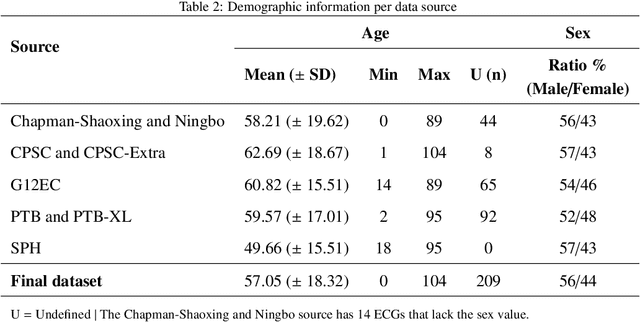
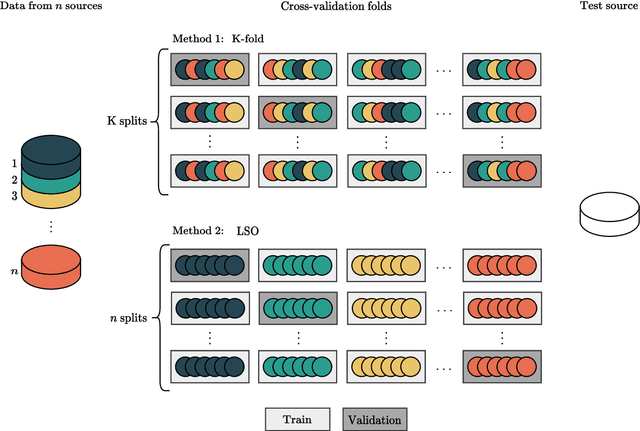
Traditionally, machine learning-based clinical prediction models have been trained and evaluated on patient data from a single source, such as a hospital. Cross-validation methods can be used to estimate the accuracy of such models on new patients originating from the same source, by repeated random splitting of the data. However, such estimates tend to be highly overoptimistic when compared to accuracy obtained from deploying models to sources not represented in the dataset, such as a new hospital. The increasing availability of multi-source medical datasets provides new opportunities for obtaining more comprehensive and realistic evaluations of expected accuracy through source-level cross-validation designs. In this study, we present a systematic empirical evaluation of standard K-fold cross-validation and leave-source-out cross-validation methods in a multi-source setting. We consider the task of electrocardiogram based cardiovascular disease classification, combining and harmonizing the openly available PhysioNet CinC Challenge 2021 and the Shandong Provincial Hospital datasets for our study. Our results show that K-fold cross-validation, both on single-source and multi-source data, systemically overestimates prediction performance when the end goal is to generalize to new sources. Leave-source-out cross-validation provides more reliable performance estimates, having close to zero bias though larger variability. The evaluation highlights the dangers of obtaining misleading cross-validation results on medical data and demonstrates how these issues can be mitigated when having access to multi-source data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge