Bernard De Baets
A Comparative Survey of Vision Transformers for Feature Extraction in Texture Analysis
Jun 10, 2024



Abstract:Texture, a significant visual attribute in images, has been extensively investigated across various image recognition applications. Convolutional Neural Networks (CNNs), which have been successful in many computer vision tasks, are currently among the best texture analysis approaches. On the other hand, Vision Transformers (ViTs) have been surpassing the performance of CNNs on tasks such as object recognition, causing a paradigm shift in the field. However, ViTs have so far not been scrutinized for texture recognition, hindering a proper appreciation of their potential in this specific setting. For this reason, this work explores various pre-trained ViT architectures when transferred to tasks that rely on textures. We review 21 different ViT variants and perform an extensive evaluation and comparison with CNNs and hand-engineered models on several tasks, such as assessing robustness to changes in texture rotation, scale, and illumination, and distinguishing color textures, material textures, and texture attributes. The goal is to understand the potential and differences among these models when directly applied to texture recognition, using pre-trained ViTs primarily for feature extraction and employing linear classifiers for evaluation. We also evaluate their efficiency, which is one of the main drawbacks in contrast to other methods. Our results show that ViTs generally outperform both CNNs and hand-engineered models, especially when using stronger pre-training and tasks involving in-the-wild textures (images from the internet). We highlight the following promising models: ViT-B with DINO pre-training, BeiTv2, and the Swin architecture, as well as the EfficientFormer as a low-cost alternative. In terms of efficiency, although having a higher number of GFLOPs and parameters, ViT-B and BeiT(v2) can achieve a lower feature extraction time on GPUs compared to ResNet50.
Advanced wood species identification based on multiple anatomical sections and using deep feature transfer and fusion
Apr 12, 2024Abstract:In recent years, we have seen many advancements in wood species identification. Methods like DNA analysis, Near Infrared (NIR) spectroscopy, and Direct Analysis in Real Time (DART) mass spectrometry complement the long-established wood anatomical assessment of cell and tissue morphology. However, most of these methods have some limitations such as high costs, the need for skilled experts for data interpretation, and the lack of good datasets for professional reference. Therefore, most of these methods, and certainly the wood anatomical assessment, may benefit from tools based on Artificial Intelligence. In this paper, we apply two transfer learning techniques with Convolutional Neural Networks (CNNs) to a multi-view Congolese wood species dataset including sections from different orientations and viewed at different microscopic magnifications. We explore two feature extraction methods in detail, namely Global Average Pooling (GAP) and Random Encoding of Aggregated Deep Activation Maps (RADAM), for efficient and accurate wood species identification. Our results indicate superior accuracy on diverse datasets and anatomical sections, surpassing the results of other methods. Our proposal represents a significant advancement in wood species identification, offering a robust tool to support the conservation of forest ecosystems and promote sustainable forestry practices.
Hyperdimensional computing: a fast, robust and interpretable paradigm for biological data
Feb 27, 2024Abstract:Advances in bioinformatics are primarily due to new algorithms for processing diverse biological data sources. While sophisticated alignment algorithms have been pivotal in analyzing biological sequences, deep learning has substantially transformed bioinformatics, addressing sequence, structure, and functional analyses. However, these methods are incredibly data-hungry, compute-intensive and hard to interpret. Hyperdimensional computing (HDC) has recently emerged as an intriguing alternative. The key idea is that random vectors of high dimensionality can represent concepts such as sequence identity or phylogeny. These vectors can then be combined using simple operators for learning, reasoning or querying by exploiting the peculiar properties of high-dimensional spaces. Our work reviews and explores the potential of HDC for bioinformatics, emphasizing its efficiency, interpretability, and adeptness in handling multimodal and structured data. HDC holds a lot of potential for various omics data searching, biosignal analysis and health applications.
Prediction of Activated Sludge Settling Characteristics from Microscopy Images with Deep Convolutional Neural Networks and Transfer Learning
Feb 14, 2024Abstract:Microbial communities play a key role in biological wastewater treatment processes. Activated sludge settling characteristics, for example, are affected by microbial community composition, varying by changes in operating conditions and influent characteristics of wastewater treatment plants (WWTPs). Timely assessment and prediction of changes in microbial composition leading to settling problems, such as filamentous bulking (FB), can prevent operational challenges, reductions in treatment efficiency, and adverse environmental impacts. This study presents an innovative computer vision-based approach to assess activated sludge-settling characteristics based on the morphological properties of flocs and filaments in microscopy images. Implementing the transfer learning of deep convolutional neural network (CNN) models, this approach aims to overcome the limitations of existing quantitative image analysis techniques. The offline microscopy image dataset was collected over two years, with weekly sampling at a full-scale industrial WWTP in Belgium. Multiple data augmentation techniques were employed to enhance the generalizability of the CNN models. Various CNN architectures, including Inception v3, ResNet18, ResNet152, ConvNeXt-nano, and ConvNeXt-S, were tested to evaluate their performance in predicting sludge settling characteristics. The sludge volume index was used as the final prediction variable, but the method can easily be adjusted to predict any other settling metric of choice. The results showed that the suggested CNN-based approach provides less labour-intensive, objective, and consistent assessments, while transfer learning notably minimises the training phase, resulting in a generalizable system that can be employed in real-time applications.
The Hyperdimensional Transform for Distributional Modelling, Regression and Classification
Nov 14, 2023Abstract:Hyperdimensional computing (HDC) is an increasingly popular computing paradigm with immense potential for future intelligent applications. Although the main ideas already took form in the 1990s, HDC recently gained significant attention, especially in the field of machine learning and data science. Next to efficiency, interoperability and explainability, HDC offers attractive properties for generalization as it can be seen as an attempt to combine connectionist ideas from neural networks with symbolic aspects. In recent work, we introduced the hyperdimensional transform, revealing deep theoretical foundations for representing functions and distributions as high-dimensional holographic vectors. Here, we present the power of the hyperdimensional transform to a broad data science audience. We use the hyperdimensional transform as a theoretical basis and provide insight into state-of-the-art HDC approaches for machine learning. We show how existing algorithms can be modified and how this transform can lead to a novel, well-founded toolbox. Next to the standard regression and classification tasks of machine learning, our discussion includes various aspects of statistical modelling, such as representation, learning and deconvolving distributions, sampling, Bayesian inference, and uncertainty estimation.
The Hyperdimensional Transform: a Holographic Representation of Functions
Oct 24, 2023Abstract:Integral transforms are invaluable mathematical tools to map functions into spaces where they are easier to characterize. We introduce the hyperdimensional transform as a new kind of integral transform. It converts square-integrable functions into noise-robust, holographic, high-dimensional representations called hyperdimensional vectors. The central idea is to approximate a function by a linear combination of random functions. We formally introduce a set of stochastic, orthogonal basis functions and define the hyperdimensional transform and its inverse. We discuss general transform-related properties such as its uniqueness, approximation properties of the inverse transform, and the representation of integrals and derivatives. The hyperdimensional transform offers a powerful, flexible framework that connects closely with other integral transforms, such as the Fourier, Laplace, and fuzzy transforms. Moreover, it provides theoretical foundations and new insights for the field of hyperdimensional computing, a computing paradigm that is rapidly gaining attention for efficient and explainable machine learning algorithms, with potential applications in statistical modelling and machine learning. In addition, we provide straightforward and easily understandable code, which can function as a tutorial and allows for the reproduction of the demonstrated examples, from computing the transform to solving differential equations.
Heteroskedastic conformal regression
Sep 15, 2023



Abstract:Conformal prediction, and split conformal prediction as a specific implementation, offer a distribution-free approach to estimating prediction intervals with statistical guarantees. Recent work has shown that split conformal prediction can produce state-of-the-art prediction intervals when focusing on marginal coverage, i.e., on a calibration dataset the method produces on average prediction intervals that contain the ground truth with a predefined coverage level. However, such intervals are often not adaptive, which can be problematic for regression problems with heteroskedastic noise. This paper tries to shed new light on how adaptive prediction intervals can be constructed using methods such as normalized and Mondrian conformal prediction. We present theoretical and experimental results in which these methods are investigated in a systematic way.
RADAM: Texture Recognition through Randomized Aggregated Encoding of Deep Activation Maps
Mar 08, 2023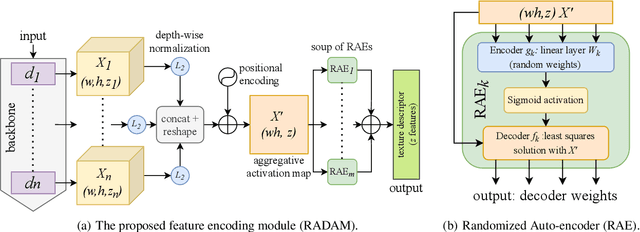
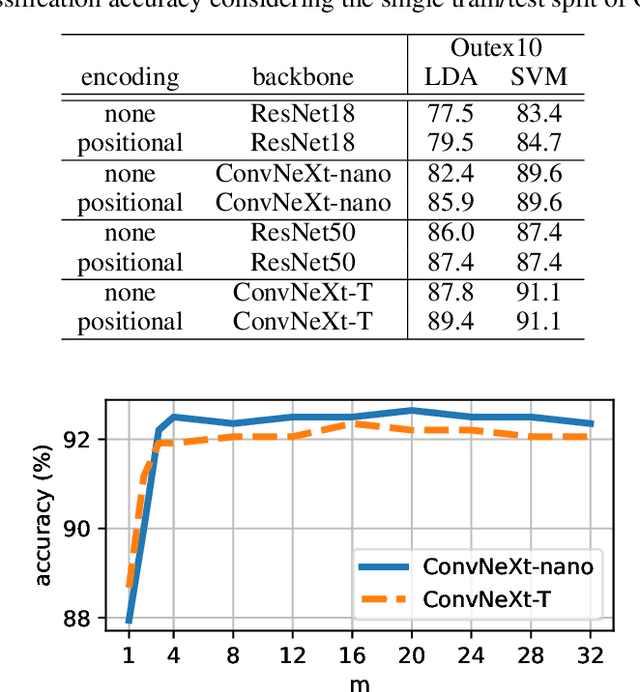
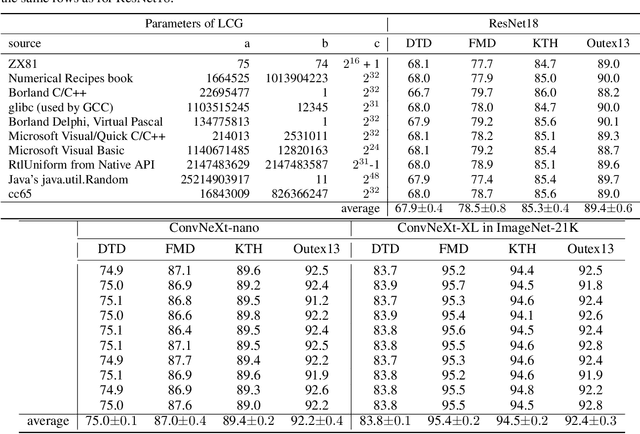
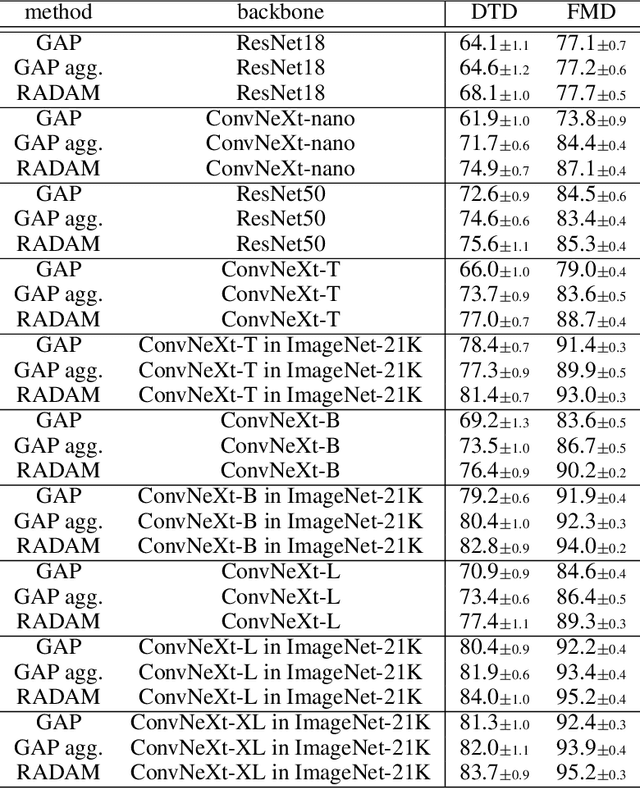
Abstract:Texture analysis is a classical yet challenging task in computer vision for which deep neural networks are actively being applied. Most approaches are based on building feature aggregation modules around a pre-trained backbone and then fine-tuning the new architecture on specific texture recognition tasks. Here we propose a new method named \textbf{R}andom encoding of \textbf{A}ggregated \textbf{D}eep \textbf{A}ctivation \textbf{M}aps (RADAM) which extracts rich texture representations without ever changing the backbone. The technique consists of encoding the output at different depths of a pre-trained deep convolutional network using a Randomized Autoencoder (RAE). The RAE is trained locally to each image using a closed-form solution, and its decoder weights are used to compose a 1-dimensional texture representation that is fed into a linear SVM. This means that no fine-tuning or backpropagation is needed. We explore RADAM on several texture benchmarks and achieve state-of-the-art results with different computational budgets. Our results suggest that pre-trained backbones may not require additional fine-tuning for texture recognition if their learned representations are better encoded.
Hyperparameter optimization in deep multi-target prediction
Nov 08, 2022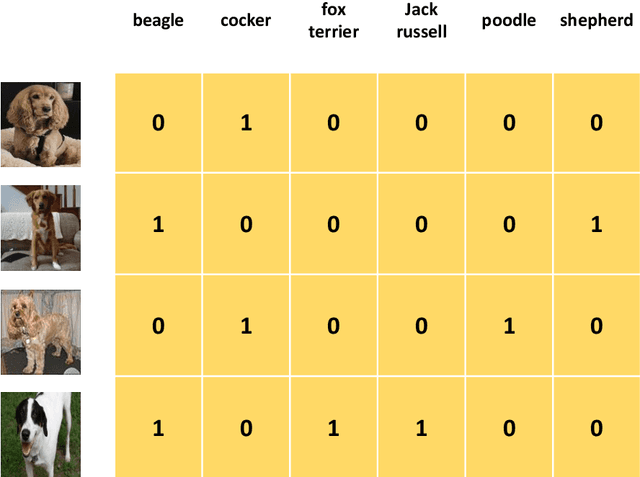
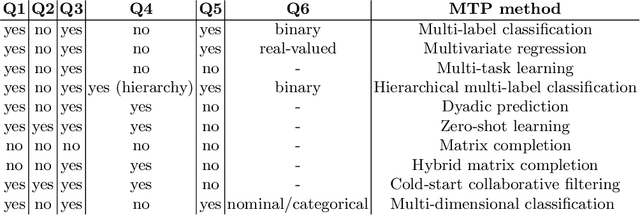
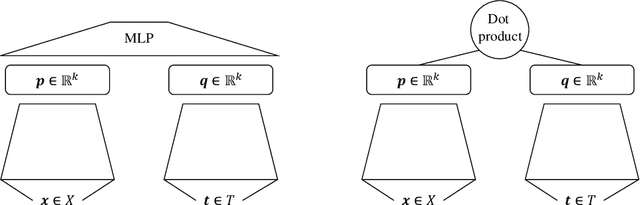
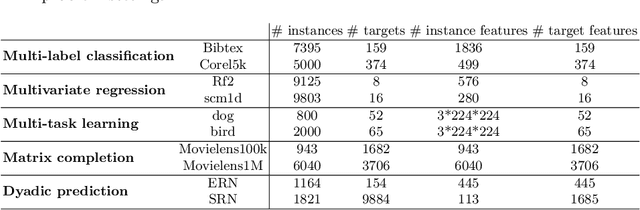
Abstract:As a result of the ever increasing complexity of configuring and fine-tuning machine learning models, the field of automated machine learning (AutoML) has emerged over the past decade. However, software implementations like Auto-WEKA and Auto-sklearn typically focus on classical machine learning (ML) tasks such as classification and regression. Our work can be seen as the first attempt at offering a single AutoML framework for most problem settings that fall under the umbrella of multi-target prediction, which includes popular ML settings such as multi-label classification, multivariate regression, multi-task learning, dyadic prediction, matrix completion, and zero-shot learning. Automated problem selection and model configuration are achieved by extending DeepMTP, a general deep learning framework for MTP problem settings, with popular hyperparameter optimization (HPO) methods. Our extensive benchmarking across different datasets and MTP problem settings identifies cases where specific HPO methods outperform others.
Improving Deep Neural Network Random Initialization Through Neuronal Rewiring
Jul 17, 2022



Abstract:The deep learning literature is continuously updated with new architectures and training techniques. However, weight initialization is overlooked by most recent research, despite some intriguing findings regarding random weights. On the other hand, recent works have been approaching Network Science to understand the structure and dynamics of Artificial Neural Networks (ANNs) after training. Therefore, in this work, we analyze the centrality of neurons in randomly initialized networks. We show that a higher neuronal strength variance may decrease performance, while a lower neuronal strength variance usually improves it. A new method is then proposed to rewire neuronal connections according to a preferential attachment (PA) rule based on their strength, which significantly reduces the strength variance of layers initialized by common methods. In this sense, PA rewiring only reorganizes connections, while preserving the magnitude and distribution of the weights. We show through an extensive statistical analysis in image classification that performance is improved in most cases, both during training and testing, when using both simple and complex architectures and learning schedules. Our results show that, aside from the magnitude, the organization of the weights is also relevant for better initialization of deep ANNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge