Gisele Miranda
Prediction of Activated Sludge Settling Characteristics from Microscopy Images with Deep Convolutional Neural Networks and Transfer Learning
Feb 14, 2024Abstract:Microbial communities play a key role in biological wastewater treatment processes. Activated sludge settling characteristics, for example, are affected by microbial community composition, varying by changes in operating conditions and influent characteristics of wastewater treatment plants (WWTPs). Timely assessment and prediction of changes in microbial composition leading to settling problems, such as filamentous bulking (FB), can prevent operational challenges, reductions in treatment efficiency, and adverse environmental impacts. This study presents an innovative computer vision-based approach to assess activated sludge-settling characteristics based on the morphological properties of flocs and filaments in microscopy images. Implementing the transfer learning of deep convolutional neural network (CNN) models, this approach aims to overcome the limitations of existing quantitative image analysis techniques. The offline microscopy image dataset was collected over two years, with weekly sampling at a full-scale industrial WWTP in Belgium. Multiple data augmentation techniques were employed to enhance the generalizability of the CNN models. Various CNN architectures, including Inception v3, ResNet18, ResNet152, ConvNeXt-nano, and ConvNeXt-S, were tested to evaluate their performance in predicting sludge settling characteristics. The sludge volume index was used as the final prediction variable, but the method can easily be adjusted to predict any other settling metric of choice. The results showed that the suggested CNN-based approach provides less labour-intensive, objective, and consistent assessments, while transfer learning notably minimises the training phase, resulting in a generalizable system that can be employed in real-time applications.
Adding Seemingly Uninformative Labels Helps in Low Data Regimes
Aug 11, 2020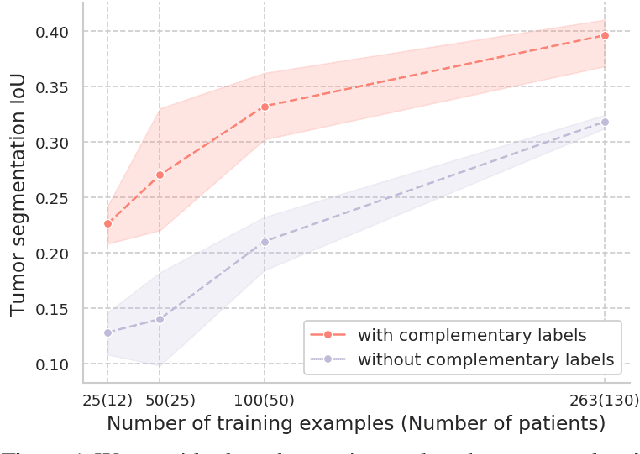
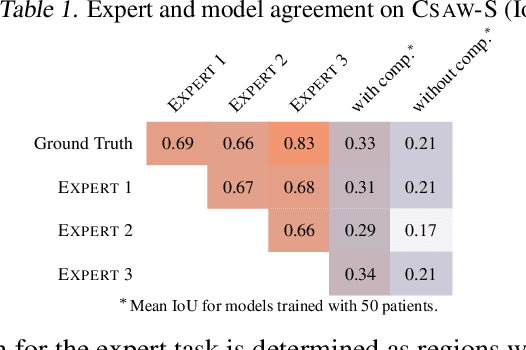
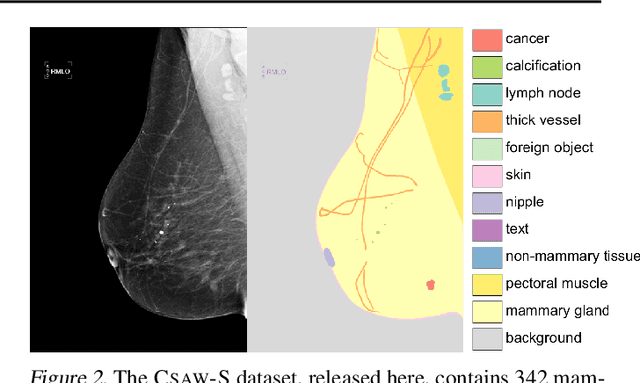
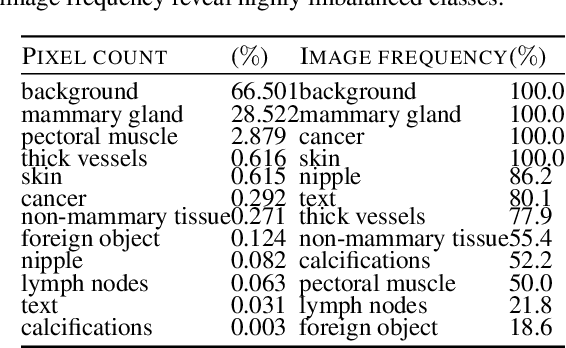
Abstract:Evidence suggests that networks trained on large datasets generalize well not solely because of the numerous training examples, but also class diversity which encourages learning of enriched features. This raises the question of whether this remains true when data is scarce - is there an advantage to learning with additional labels in low-data regimes? In this work, we consider a task that requires difficult-to-obtain expert annotations: tumor segmentation in mammography images. We show that, in low-data settings, performance can be improved by complementing the expert annotations with seemingly uninformative labels from non-expert annotators, turning the task into a multi-class problem. We reveal that these gains increase when less expert data is available, and uncover several interesting properties through further studies. We demonstrate our findings on CSAW-S, a new dataset that we introduce here, and confirm them on two public datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge