Johan Fredin Haslum
Bridging Generalization Gaps in High Content Imaging Through Online Self-Supervised Domain Adaptation
Nov 21, 2023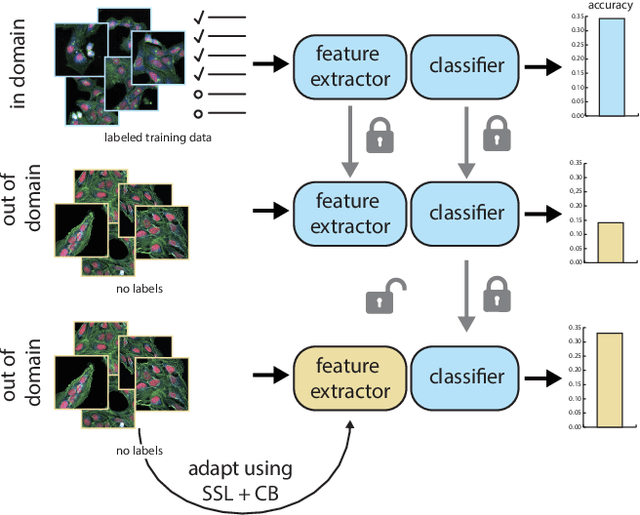
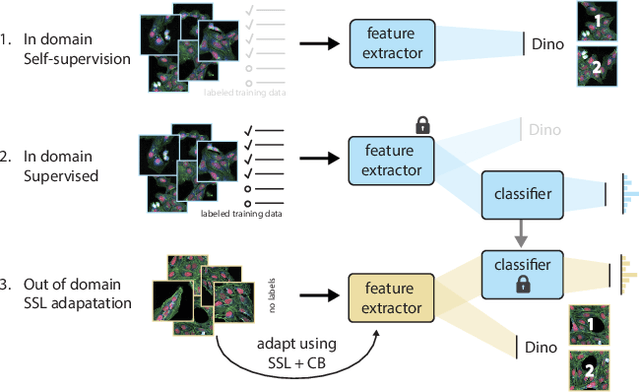
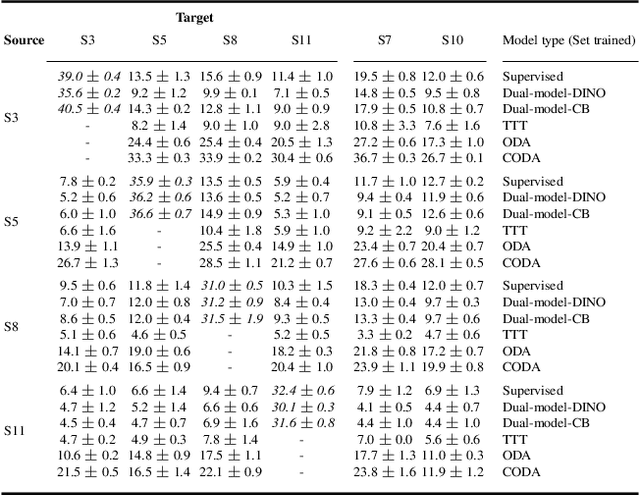
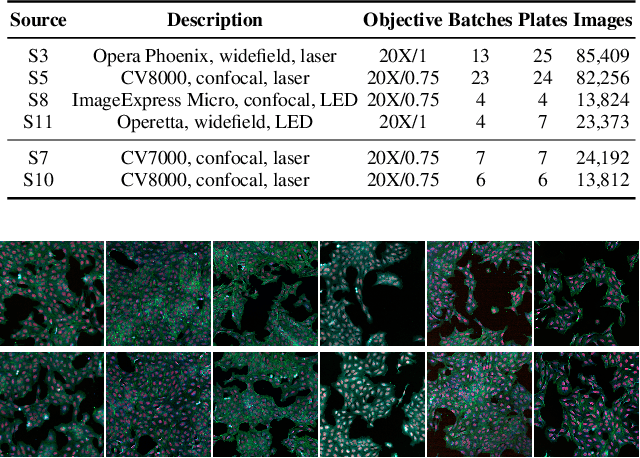
Abstract:High Content Imaging (HCI) plays a vital role in modern drug discovery and development pipelines, facilitating various stages from hit identification to candidate drug characterization. Applying machine learning models to these datasets can prove challenging as they typically consist of multiple batches, affected by experimental variation, especially if different imaging equipment have been used. Moreover, as new data arrive, it is preferable that they are analyzed in an online fashion. To overcome this, we propose CODA, an online self-supervised domain adaptation approach. CODA divides the classifier's role into a generic feature extractor and a task-specific model. We adapt the feature extractor's weights to the new domain using cross-batch self-supervision while keeping the task-specific model unchanged. Our results demonstrate that this strategy significantly reduces the generalization gap, achieving up to a 300% improvement when applied to data from different labs utilizing different microscopes. CODA can be applied to new, unlabeled out-of-domain data sources of different sizes, from a single plate to multiple experimental batches.
Are Natural Domain Foundation Models Useful for Medical Image Classification?
Oct 30, 2023
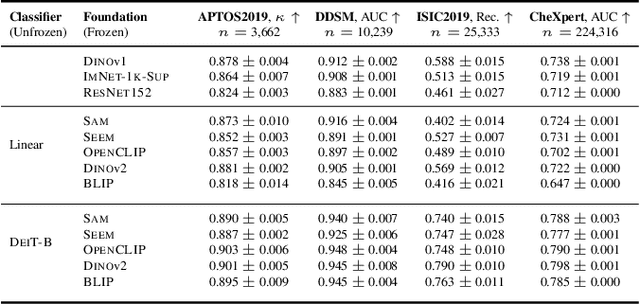
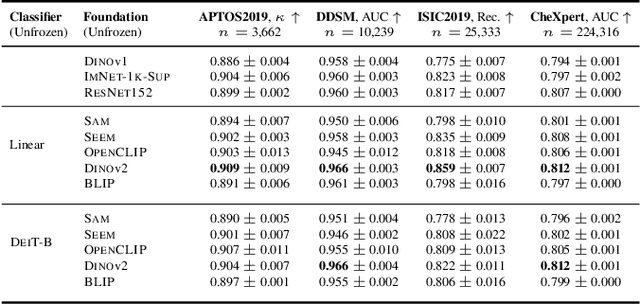

Abstract:The deep learning field is converging towards the use of general foundation models that can be easily adapted for diverse tasks. While this paradigm shift has become common practice within the field of natural language processing, progress has been slower in computer vision. In this paper we attempt to address this issue by investigating the transferability of various state-of-the-art foundation models to medical image classification tasks. Specifically, we evaluate the performance of five foundation models, namely SAM, SEEM, DINOv2, BLIP, and OpenCLIP across four well-established medical imaging datasets. We explore different training settings to fully harness the potential of these models. Our study shows mixed results. DINOv2 in particular, consistently outperforms the standard practice of ImageNet pretraining. However, other foundation models failed to consistently beat this established baseline indicating limitations in their transferability to medical image classification tasks.
Pretrained ViTs Yield Versatile Representations For Medical Images
Mar 14, 2023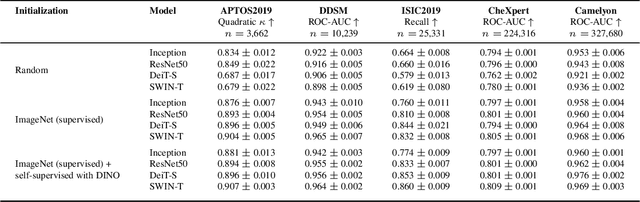
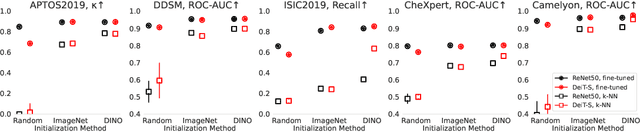
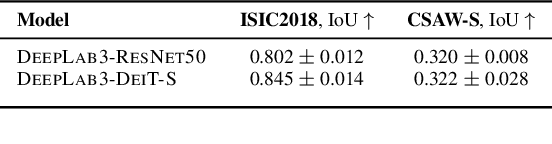
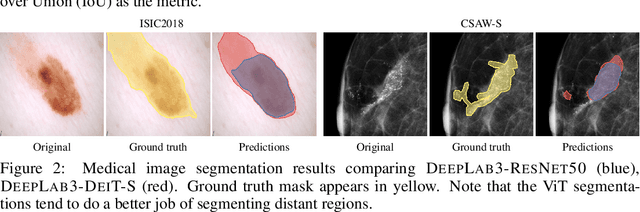
Abstract:Convolutional Neural Networks (CNNs) have reigned for a decade as the de facto approach to automated medical image diagnosis, pushing the state-of-the-art in classification, detection and segmentation tasks. Over the last years, vision transformers (ViTs) have appeared as a competitive alternative to CNNs, yielding impressive levels of performance in the natural image domain, while possessing several interesting properties that could prove beneficial for medical imaging tasks. In this work, we explore the benefits and drawbacks of transformer-based models for medical image classification. We conduct a series of experiments on several standard 2D medical image benchmark datasets and tasks. Our findings show that, while CNNs perform better if trained from scratch, off-the-shelf vision transformers can perform on par with CNNs when pretrained on ImageNet, both in a supervised and self-supervised setting, rendering them as a viable alternative to CNNs.
Metadata-guided Consistency Learning for High Content Images
Dec 22, 2022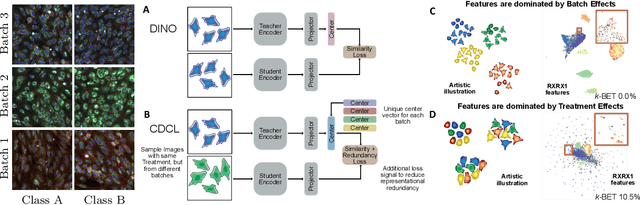
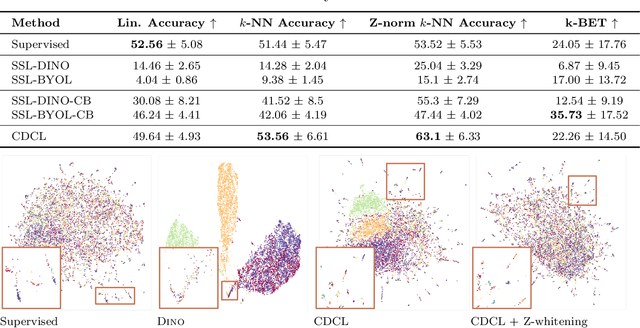

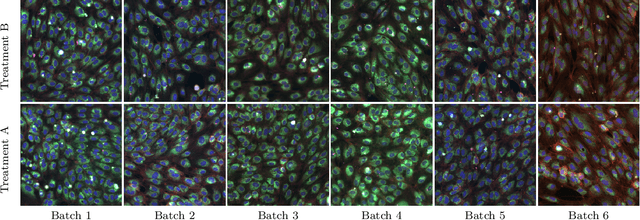
Abstract:High content imaging assays can capture rich phenotypic response data for large sets of compound treatments, aiding in the characterization and discovery of novel drugs. However, extracting representative features from high content images that can capture subtle nuances in phenotypes remains challenging. The lack of high-quality labels makes it difficult to achieve satisfactory results with supervised deep learning. Self-Supervised learning methods, which learn from automatically generated labels has shown great success on natural images, offer an attractive alternative also to microscopy images. However, we find that self-supervised learning techniques underperform on high content imaging assays. One challenge is the undesirable domain shifts present in the data known as batch effects, which may be caused by biological noise or uncontrolled experimental conditions. To this end, we introduce Cross-Domain Consistency Learning (CDCL), a novel approach that is able to learn in the presence of batch effects. CDCL enforces the learning of biological similarities while disregarding undesirable batch-specific signals, which leads to more useful and versatile representations. These features are organised according to their morphological changes and are more useful for downstream tasks - such as distinguishing treatments and mode of action.
What Makes Transfer Learning Work For Medical Images: Feature Reuse & Other Factors
Mar 02, 2022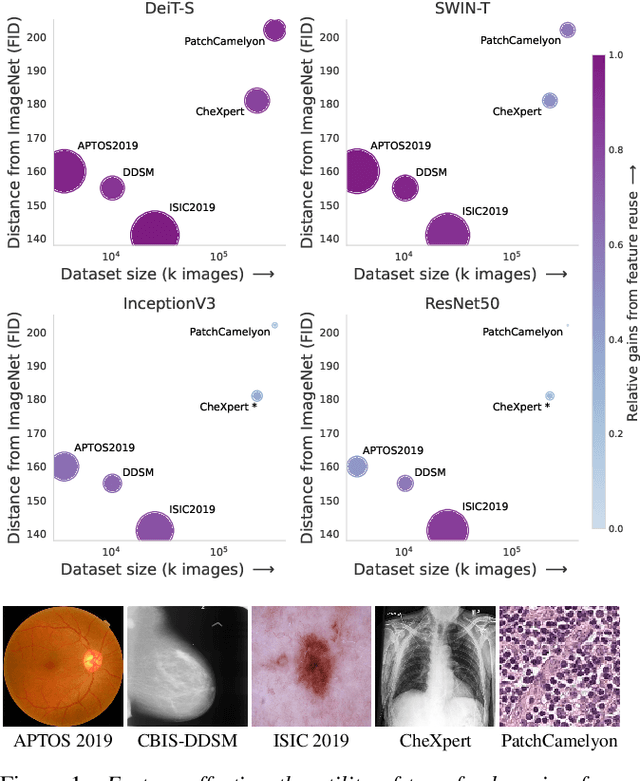
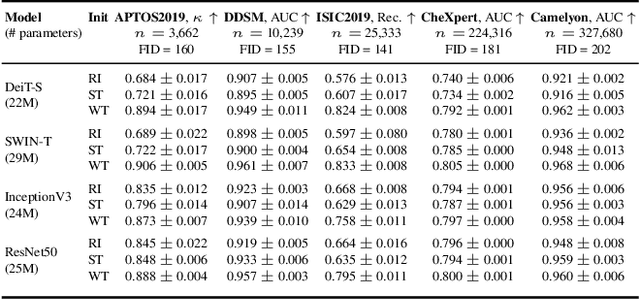
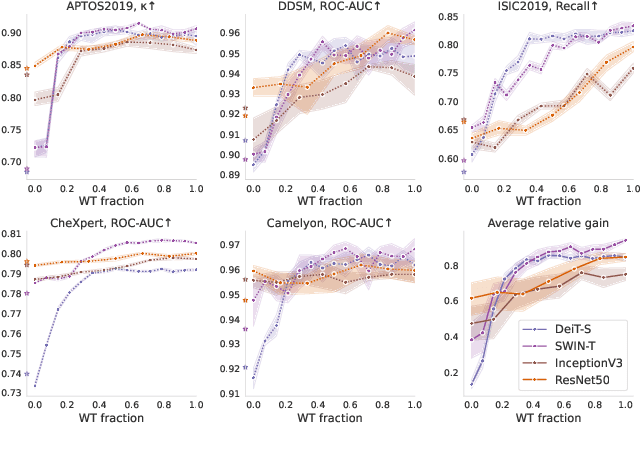
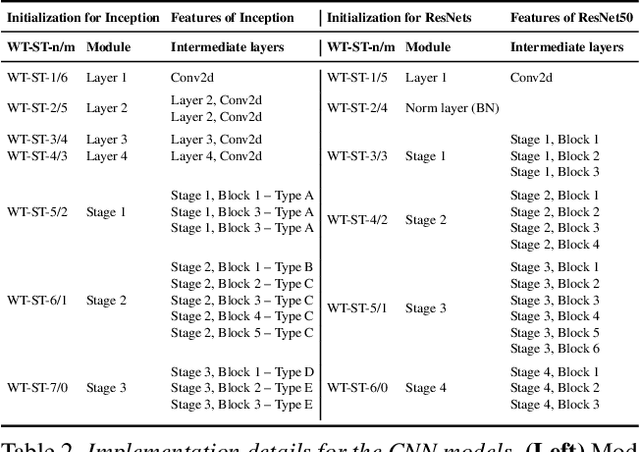
Abstract:Transfer learning is a standard technique to transfer knowledge from one domain to another. For applications in medical imaging, transfer from ImageNet has become the de-facto approach, despite differences in the tasks and image characteristics between the domains. However, it is unclear what factors determine whether - and to what extent - transfer learning to the medical domain is useful. The long-standing assumption that features from the source domain get reused has recently been called into question. Through a series of experiments on several medical image benchmark datasets, we explore the relationship between transfer learning, data size, the capacity and inductive bias of the model, as well as the distance between the source and target domain. Our findings suggest that transfer learning is beneficial in most cases, and we characterize the important role feature reuse plays in its success.
Is it Time to Replace CNNs with Transformers for Medical Images?
Aug 20, 2021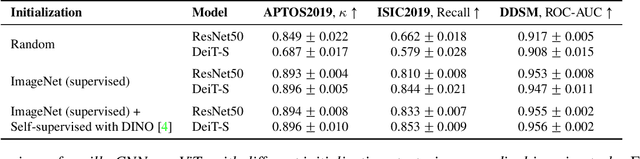
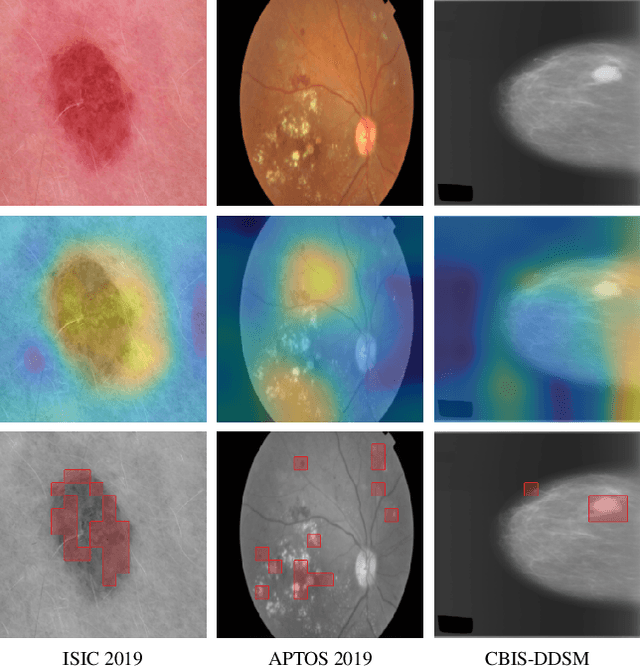
Abstract:Convolutional Neural Networks (CNNs) have reigned for a decade as the de facto approach to automated medical image diagnosis. Recently, vision transformers (ViTs) have appeared as a competitive alternative to CNNs, yielding similar levels of performance while possessing several interesting properties that could prove beneficial for medical imaging tasks. In this work, we explore whether it is time to move to transformer-based models or if we should keep working with CNNs - can we trivially switch to transformers? If so, what are the advantages and drawbacks of switching to ViTs for medical image diagnosis? We consider these questions in a series of experiments on three mainstream medical image datasets. Our findings show that, while CNNs perform better when trained from scratch, off-the-shelf vision transformers using default hyperparameters are on par with CNNs when pretrained on ImageNet, and outperform their CNN counterparts when pretrained using self-supervision.
Adding Seemingly Uninformative Labels Helps in Low Data Regimes
Aug 11, 2020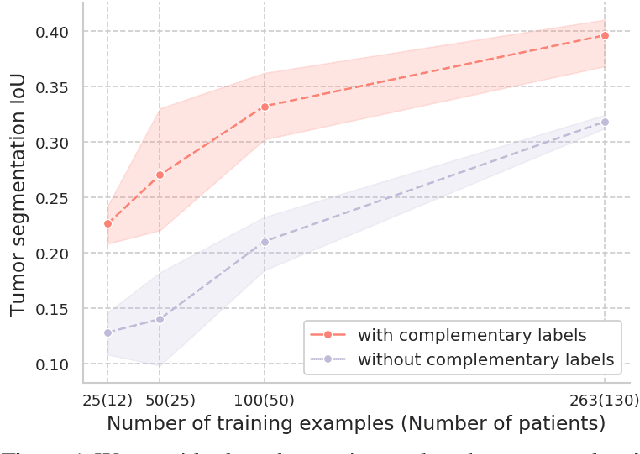
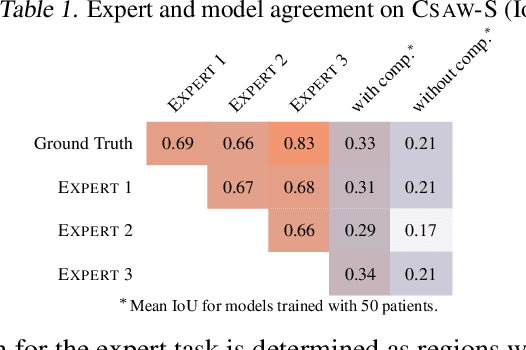
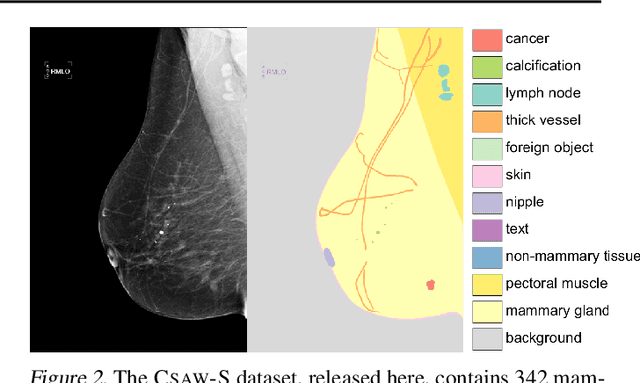
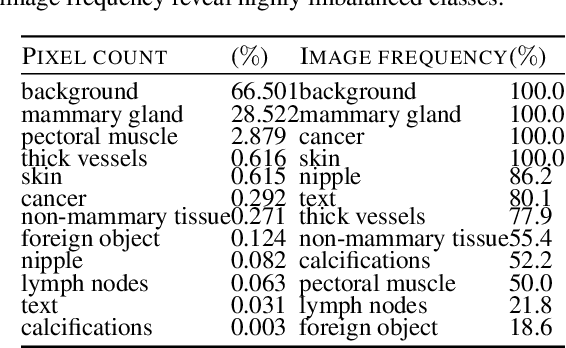
Abstract:Evidence suggests that networks trained on large datasets generalize well not solely because of the numerous training examples, but also class diversity which encourages learning of enriched features. This raises the question of whether this remains true when data is scarce - is there an advantage to learning with additional labels in low-data regimes? In this work, we consider a task that requires difficult-to-obtain expert annotations: tumor segmentation in mammography images. We show that, in low-data settings, performance can be improved by complementing the expert annotations with seemingly uninformative labels from non-expert annotators, turning the task into a multi-class problem. We reveal that these gains increase when less expert data is available, and uncover several interesting properties through further studies. We demonstrate our findings on CSAW-S, a new dataset that we introduce here, and confirm them on two public datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge