Kevin Smith
Massachusetts Institute of Technology
"Just in Time" World Modeling Supports Human Planning and Reasoning
Jan 20, 2026Abstract:Probabilistic mental simulation is thought to play a key role in human reasoning, planning, and prediction, yet the demands of simulation in complex environments exceed realistic human capacity limits. A theory with growing evidence is that people simulate using simplified representations of the environment that abstract away from irrelevant details, but it is unclear how people determine these simplifications efficiently. Here, we present a "Just-in-Time" framework for simulation-based reasoning that demonstrates how such representations can be constructed online with minimal added computation. The model uses a tight interleaving of simulation, visual search, and representation modification, with the current simulation guiding where to look and visual search flagging objects that should be encoded for subsequent simulation. Despite only ever encoding a small subset of objects, the model makes high-utility predictions. We find strong empirical support for this account over alternative models in a grid-world planning task and a physical reasoning task across a range of behavioral measures. Together, these results offer a concrete algorithmic account of how people construct reduced representations to support efficient mental simulation.
Efficient Self-Supervised Adaptation for Medical Image Analysis
Mar 24, 2025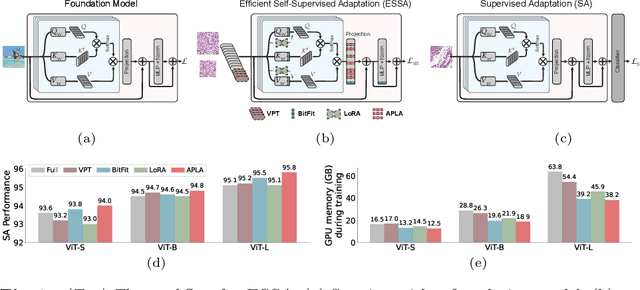
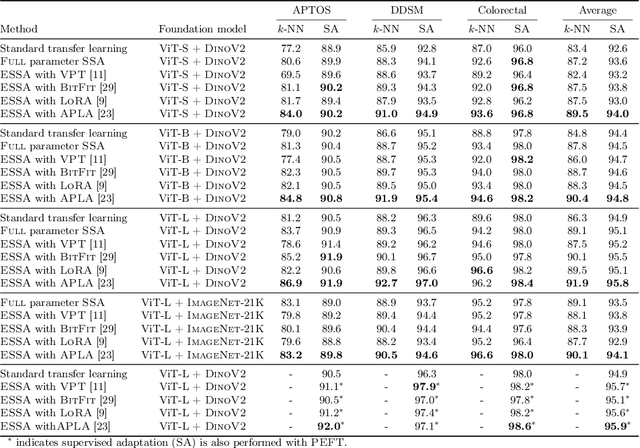
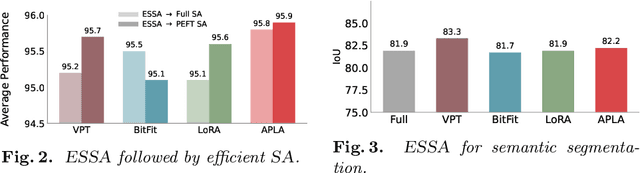
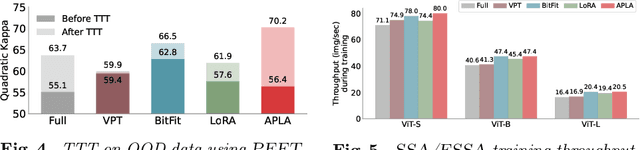
Abstract:Self-supervised adaptation (SSA) improves foundation model transfer to medical domains but is computationally prohibitive. Although parameter efficient fine-tuning methods such as LoRA have been explored for supervised adaptation, their effectiveness for SSA remains unknown. In this work, we introduce efficient self-supervised adaptation (ESSA), a framework that applies parameter-efficient fine-tuning techniques to SSA with the aim of reducing computational cost and improving adaptation performance. Among the methods tested, Attention Projection Layer Adaptation (APLA) sets a new state-of-the-art, consistently surpassing full-parameter SSA and supervised fine-tuning across diverse medical tasks, while reducing GPU memory by up to 40.1% and increasing training throughput by 25.2%, all while maintaining inference efficiency.
APLA: A Simple Adaptation Method for Vision Transformers
Mar 14, 2025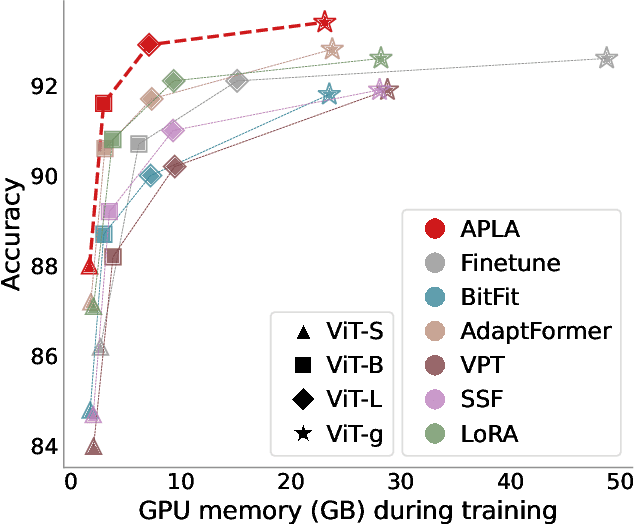
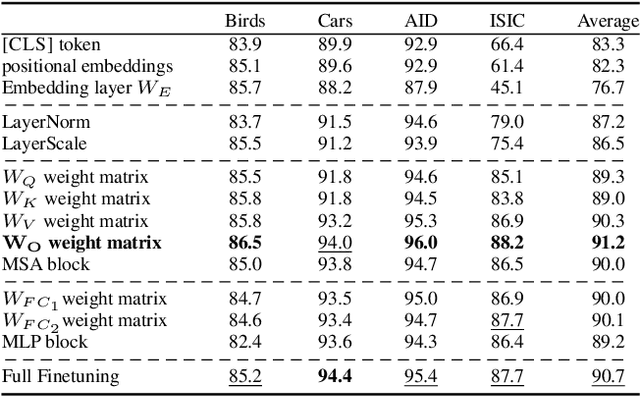
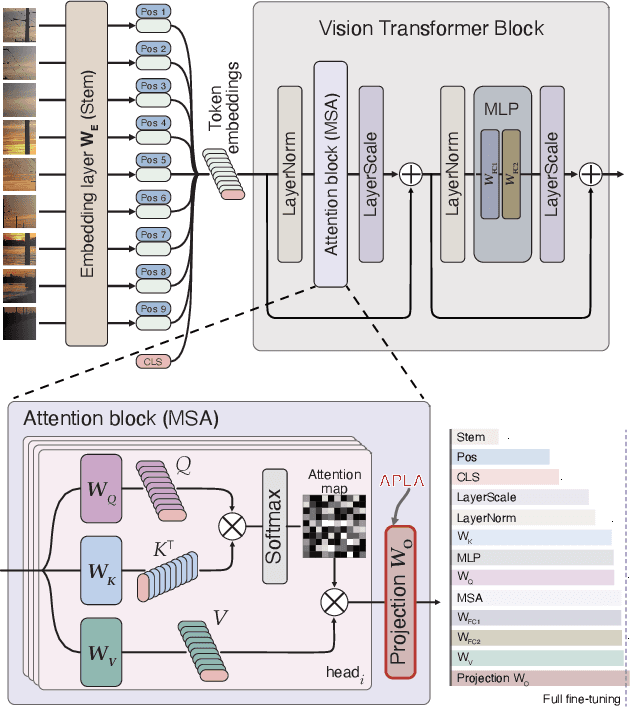
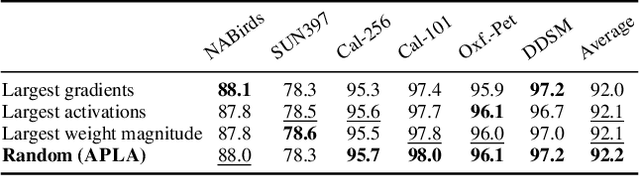
Abstract:Existing adaptation techniques typically require architectural modifications or added parameters, leading to high computational costs and complexity. We introduce Attention Projection Layer Adaptation (APLA), a simple approach to adapt vision transformers (ViTs) without altering the architecture or adding parameters. Through a systematic analysis, we find that the layer immediately after the attention mechanism is crucial for adaptation. By updating only this projection layer, or even just a random subset of this layer's weights, APLA achieves state-of-the-art performance while reducing GPU memory usage by up to 52.63% and training time by up to 43.0%, with no extra cost at inference. Across 46 datasets covering a variety of tasks including scene classification, medical imaging, satellite imaging, and fine-grained classification, APLA consistently outperforms 17 other leading adaptation methods, including full fine-tuning, on classification, segmentation, and detection tasks. The code is available at https://github.com/MoeinSorkhei/APLA.
VORTEX: Challenging CNNs at Texture Recognition by using Vision Transformers with Orderless and Randomized Token Encodings
Mar 09, 2025



Abstract:Texture recognition has recently been dominated by ImageNet-pre-trained deep Convolutional Neural Networks (CNNs), with specialized modifications and feature engineering required to achieve state-of-the-art (SOTA) performance. However, although Vision Transformers (ViTs) were introduced a few years ago, little is known about their texture recognition ability. Therefore, in this work, we introduce VORTEX (ViTs with Orderless and Randomized Token Encodings for Texture Recognition), a novel method that enables the effective use of ViTs for texture analysis. VORTEX extracts multi-depth token embeddings from pre-trained ViT backbones and employs a lightweight module to aggregate hierarchical features and perform orderless encoding, obtaining a better image representation for texture recognition tasks. This approach allows seamless integration with any ViT with the common transformer architecture. Moreover, no fine-tuning of the backbone is performed, since they are used only as frozen feature extractors, and the features are fed to a linear SVM. We evaluate VORTEX on nine diverse texture datasets, demonstrating its ability to achieve or surpass SOTA performance in a variety of texture analysis scenarios. By bridging the gap between texture recognition with CNNs and transformer-based architectures, VORTEX paves the way for adopting emerging transformer foundation models. Furthermore, VORTEX demonstrates robust computational efficiency when coupled with ViT backbones compared to CNNs with similar costs. The method implementation and experimental scripts are publicly available in our online repository.
Random Token Fusion for Multi-View Medical Diagnosis
Oct 21, 2024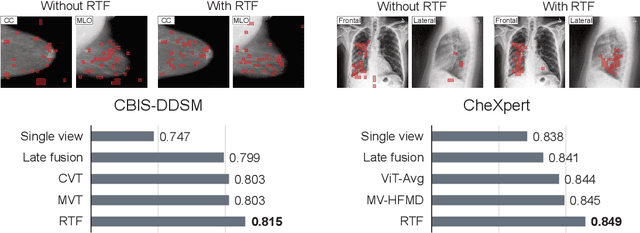

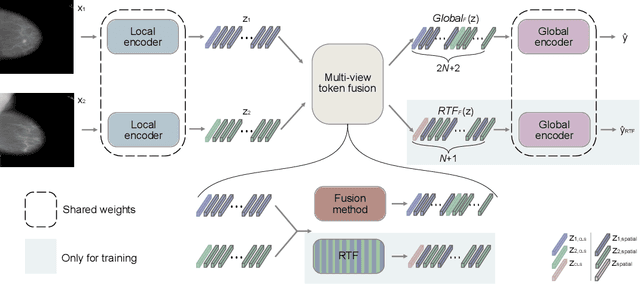

Abstract:In multi-view medical diagnosis, deep learning-based models often fuse information from different imaging perspectives to improve diagnostic performance. However, existing approaches are prone to overfitting and rely heavily on view-specific features, which can lead to trivial solutions. In this work, we introduce Random Token Fusion (RTF), a novel technique designed to enhance multi-view medical image analysis using vision transformers. By integrating randomness into the feature fusion process during training, RTF addresses the issue of overfitting and enhances the robustness and accuracy of diagnostic models without incurring any additional cost at inference. We validate our approach on standard mammography and chest X-ray benchmark datasets. Through extensive experiments, we demonstrate that RTF consistently improves the performance of existing fusion methods, paving the way for a new generation of multi-view medical foundation models.
Learning from Offline Foundation Features with Tensor Augmentations
Oct 03, 2024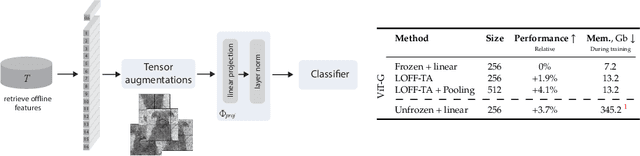
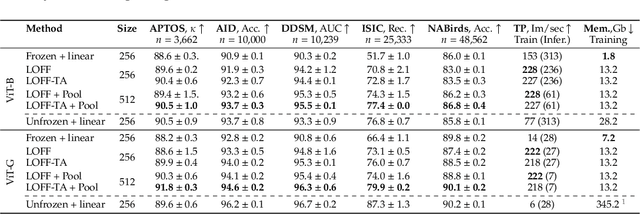
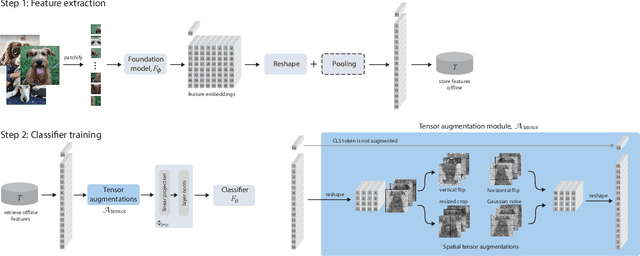
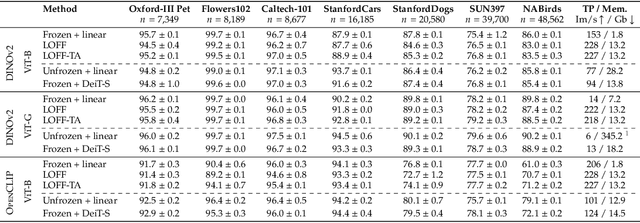
Abstract:We introduce Learning from Offline Foundation Features with Tensor Augmentations (LOFF-TA), an efficient training scheme designed to harness the capabilities of foundation models in limited resource settings where their direct development is not feasible. LOFF-TA involves training a compact classifier on cached feature embeddings from a frozen foundation model, resulting in up to $37\times$ faster training and up to $26\times$ reduced GPU memory usage. Because the embeddings of augmented images would be too numerous to store, yet the augmentation process is essential for training, we propose to apply tensor augmentations to the cached embeddings of the original non-augmented images. LOFF-TA makes it possible to leverage the power of foundation models, regardless of their size, in settings with limited computational capacity. Moreover, LOFF-TA can be used to apply foundation models to high-resolution images without increasing compute. In certain scenarios, we find that training with LOFF-TA yields better results than directly fine-tuning the foundation model.
Evaluating Large Vision-and-Language Models on Children's Mathematical Olympiads
Jun 22, 2024Abstract:Recent years have seen a significant progress in the general-purpose problem solving abilities of large vision and language models (LVLMs), such as ChatGPT, Gemini, etc.; some of these breakthroughs even seem to enable AI models to outperform human abilities in varied tasks that demand higher-order cognitive skills. Are the current large AI models indeed capable of generalized problem solving as humans do? A systematic analysis of AI capabilities for joint vision and text reasoning, however, is missing in the current scientific literature. In this paper, we make an effort towards filling this gap, by evaluating state-of-the-art LVLMs on their mathematical and algorithmic reasoning abilities using visuo-linguistic problems from children's Olympiads. Specifically, we consider problems from the Mathematical Kangaroo (MK) Olympiad, which is a popular international competition targeted at children from grades 1-12, that tests children's deeper mathematical abilities using puzzles that are appropriately gauged to their age and skills. Using the puzzles from MK, we created a dataset, dubbed SMART-840, consisting of 840 problems from years 2020-2024. With our dataset, we analyze LVLMs power on mathematical reasoning; their responses on our puzzles offer a direct way to compare against that of children. Our results show that modern LVLMs do demonstrate increasingly powerful reasoning skills in solving problems for higher grades, but lack the foundations to correctly answer problems designed for younger children. Further analysis shows that there is no significant correlation between the reasoning capabilities of AI models and that of young children, and their capabilities appear to be based on a different type of reasoning than the cumulative knowledge that underlies children's mathematics and logic skills.
MAMOC: MRI Motion Correction via Masked Autoencoding
May 23, 2024Abstract:The presence of motion artifacts in magnetic resonance imaging (MRI) scans poses a significant challenge, where even minor patient movements can lead to artifacts that may compromise the scan's utility. This paper introduces Masked Motion Correction (MAMOC), a novel method designed to address the issue of Retrospective Artifact Correction (RAC) in motion-affected MRI brain scans. MAMOC uses masked autoencoding self-supervision and test-time prediction to efficiently remove motion artifacts, producing state-of-the-art, native resolution scans. Until recently, realistic data to evaluate retrospective motion correction methods did not exist, motion artifacts had to be simulated. Leveraging the MR-ART dataset, this work is the first to evaluate motion correction in MRI scans using real motion data, showing the superiority of MAMOC to existing motion correction (MC) methods.
Bridging Generalization Gaps in High Content Imaging Through Online Self-Supervised Domain Adaptation
Nov 21, 2023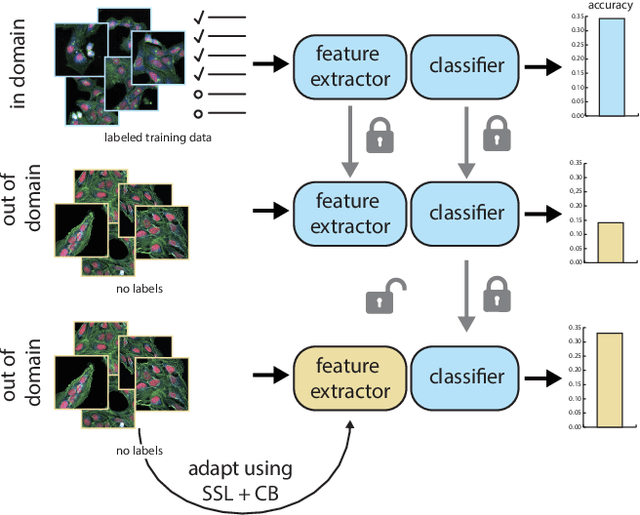
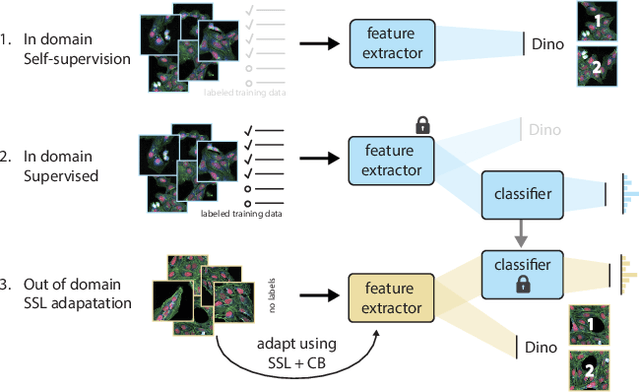
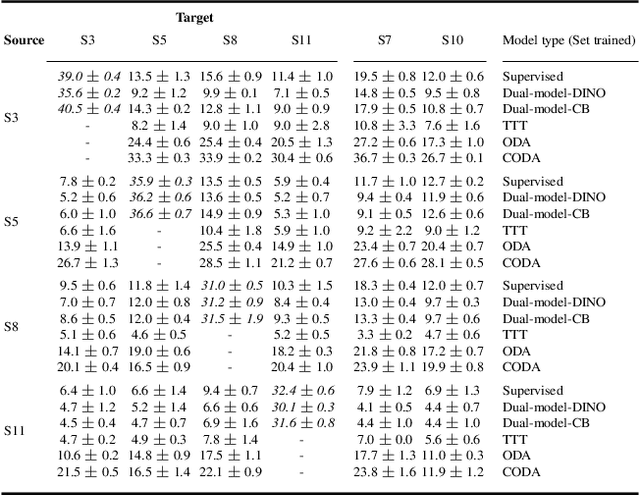
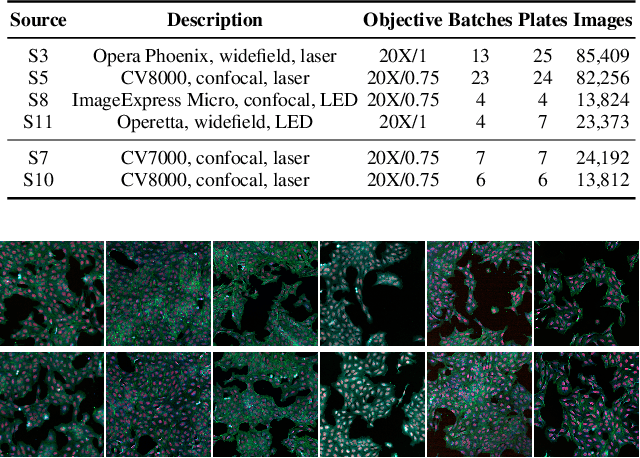
Abstract:High Content Imaging (HCI) plays a vital role in modern drug discovery and development pipelines, facilitating various stages from hit identification to candidate drug characterization. Applying machine learning models to these datasets can prove challenging as they typically consist of multiple batches, affected by experimental variation, especially if different imaging equipment have been used. Moreover, as new data arrive, it is preferable that they are analyzed in an online fashion. To overcome this, we propose CODA, an online self-supervised domain adaptation approach. CODA divides the classifier's role into a generic feature extractor and a task-specific model. We adapt the feature extractor's weights to the new domain using cross-batch self-supervision while keeping the task-specific model unchanged. Our results demonstrate that this strategy significantly reduces the generalization gap, achieving up to a 300% improvement when applied to data from different labs utilizing different microscopes. CODA can be applied to new, unlabeled out-of-domain data sources of different sizes, from a single plate to multiple experimental batches.
Are Natural Domain Foundation Models Useful for Medical Image Classification?
Oct 30, 2023
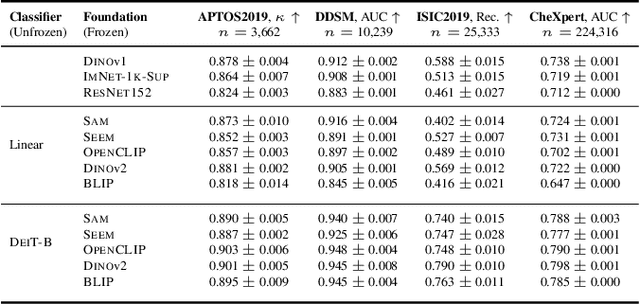
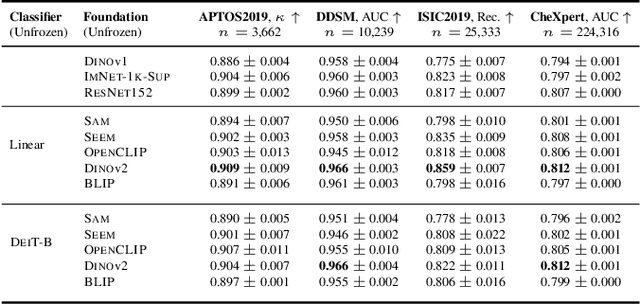

Abstract:The deep learning field is converging towards the use of general foundation models that can be easily adapted for diverse tasks. While this paradigm shift has become common practice within the field of natural language processing, progress has been slower in computer vision. In this paper we attempt to address this issue by investigating the transferability of various state-of-the-art foundation models to medical image classification tasks. Specifically, we evaluate the performance of five foundation models, namely SAM, SEEM, DINOv2, BLIP, and OpenCLIP across four well-established medical imaging datasets. We explore different training settings to fully harness the potential of these models. Our study shows mixed results. DINOv2 in particular, consistently outperforms the standard practice of ImageNet pretraining. However, other foundation models failed to consistently beat this established baseline indicating limitations in their transferability to medical image classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge