Yuan-Ting Zhang
Computation-Efficient Semi-Supervised Learning for ECG-based Cardiovascular Diseases Detection
Jun 20, 2024Abstract:Label scarcity problem is the main challenge that hinders the wide application of deep learning systems in automatic cardiovascular diseases (CVDs) detection using electrocardiography (ECG). Tuning pre-trained models alleviates this problem by transferring knowledge learned from large datasets to downstream small datasets. However, bottlenecks in computational efficiency and CVDs detection performance limit its clinical applications. It is difficult to improve the detection performance without significantly sacrificing model computational efficiency. Here, we propose a computation-efficient semi-supervised learning paradigm (FastECG) for robust and computation-efficient CVDs detection using ECG. It enables a robust adaptation of pre-trained models on downstream datasets with limited supervision and high computational efficiency. First, a random-deactivation technique is developed to achieve robust and fast low-rank adaptation of pre-trained weights. Subsequently, we propose a one-shot rank allocation module to determine the optimal ranks for the update matrices of the pre-trained weights. Finally, a lightweight semi-supervised learning pipeline is introduced to enhance model performance by leveraging labeled and unlabeled data with high computational efficiency. Extensive experiments on four downstream ECG datasets demonstrate that FastECG not only outperforms the state-of-the-art methods in multi-label CVDs detection but also consumes fewer GPU footprints, training time, and parameter storage space. As such, this paradigm provides an effective solution for achieving high computational efficiency and robust detection performance in the clinical applications of pre-trained models under limited supervision.
Semi-Supervised Learning for Multi-Label Cardiovascular Diseases Prediction:A Multi-Dataset Study
Jun 18, 2023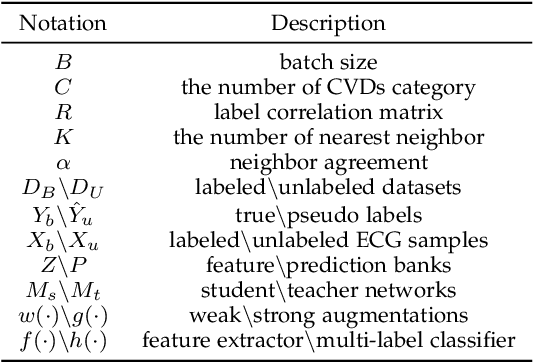
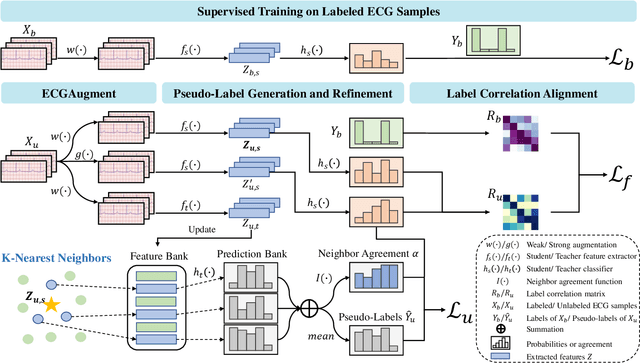
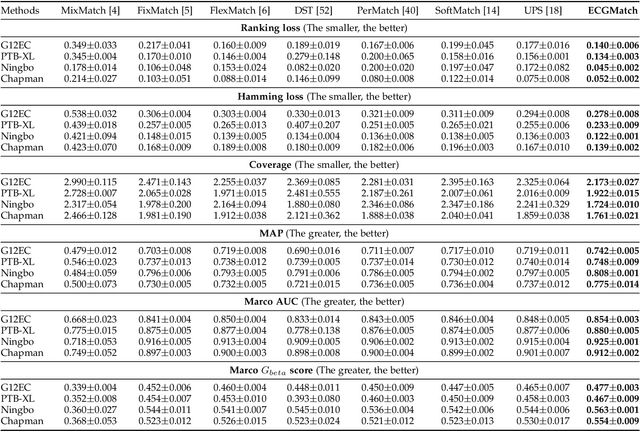
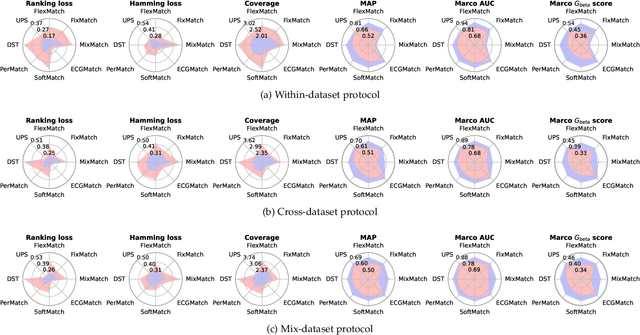
Abstract:Electrocardiography (ECG) is a non-invasive tool for predicting cardiovascular diseases (CVDs). Current ECG-based diagnosis systems show promising performance owing to the rapid development of deep learning techniques. However, the label scarcity problem, the co-occurrence of multiple CVDs and the poor performance on unseen datasets greatly hinder the widespread application of deep learning-based models. Addressing them in a unified framework remains a significant challenge. To this end, we propose a multi-label semi-supervised model (ECGMatch) to recognize multiple CVDs simultaneously with limited supervision. In the ECGMatch, an ECGAugment module is developed for weak and strong ECG data augmentation, which generates diverse samples for model training. Subsequently, a hyperparameter-efficient framework with neighbor agreement modeling and knowledge distillation is designed for pseudo-label generation and refinement, which mitigates the label scarcity problem. Finally, a label correlation alignment module is proposed to capture the co-occurrence information of different CVDs within labeled samples and propagate this information to unlabeled samples. Extensive experiments on four datasets and three protocols demonstrate the effectiveness and stability of the proposed model, especially on unseen datasets. As such, this model can pave the way for diagnostic systems that achieve robust performance on multi-label CVDs prediction with limited supervision.
EEGMatch: Learning with Incomplete Labels for Semi-Supervised EEG-based Cross-Subject Emotion Recognition
Mar 27, 2023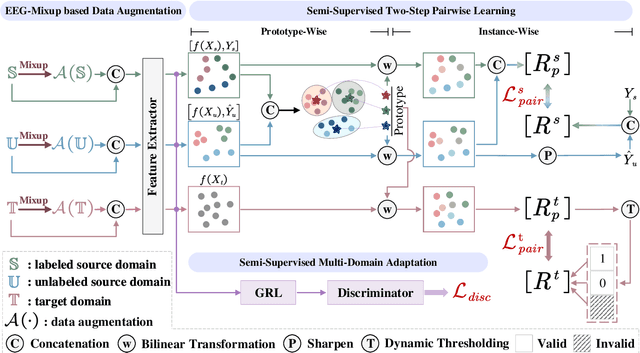
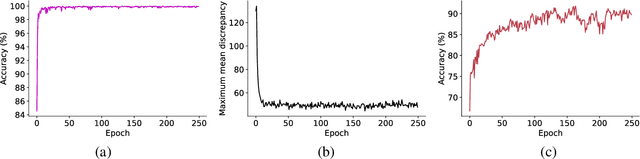
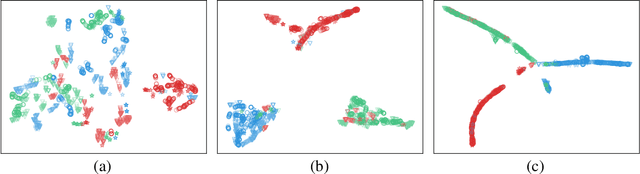
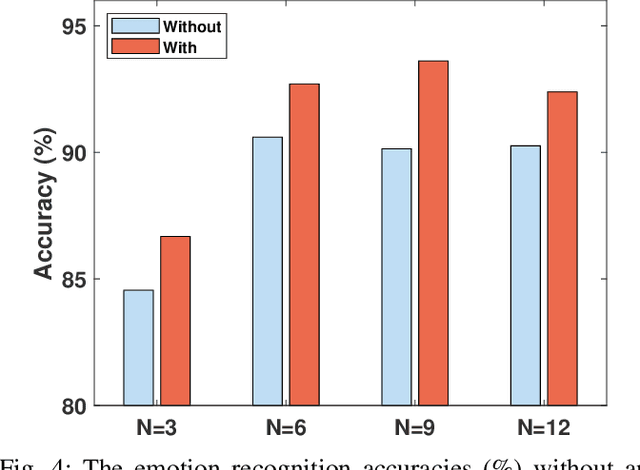
Abstract:Electroencephalography (EEG) is an objective tool for emotion recognition and shows promising performance. However, the label scarcity problem is a main challenge in this field, which limits the wide application of EEG-based emotion recognition. In this paper, we propose a novel semi-supervised learning framework (EEGMatch) to leverage both labeled and unlabeled EEG data. First, an EEG-Mixup based data augmentation method is developed to generate more valid samples for model learning. Second, a semi-supervised two-step pairwise learning method is proposed to bridge prototype-wise and instance-wise pairwise learning, where the prototype-wise pairwise learning measures the global relationship between EEG data and the prototypical representation of each emotion class and the instance-wise pairwise learning captures the local intrinsic relationship among EEG data. Third, a semi-supervised multi-domain adaptation is introduced to align the data representation among multiple domains (labeled source domain, unlabeled source domain, and target domain), where the distribution mismatch is alleviated. Extensive experiments are conducted on two benchmark databases (SEED and SEED-IV) under a cross-subject leave-one-subject-out cross-validation evaluation protocol. The results show the proposed EEGmatch performs better than the state-of-the-art methods under different incomplete label conditions (with 6.89% improvement on SEED and 1.44% improvement on SEED-IV), which demonstrates the effectiveness of the proposed EEGMatch in dealing with the label scarcity problem in emotion recognition using EEG signals. The source code is available at https://github.com/KAZABANA/EEGMatch.
Long-term Blood Pressure Prediction with Deep Recurrent Neural Networks
Jan 14, 2018



Abstract:Existing methods for arterial blood pressure (BP) estimation directly map the input physiological signals to output BP values without explicitly modeling the underlying temporal dependencies in BP dynamics. As a result, these models suffer from accuracy decay over a long time and thus require frequent calibration. In this work, we address this issue by formulating BP estimation as a sequence prediction problem in which both the input and target are temporal sequences. We propose a novel deep recurrent neural network (RNN) consisting of multilayered Long Short-Term Memory (LSTM) networks, which are incorporated with (1) a bidirectional structure to access larger-scale context information of input sequence, and (2) residual connections to allow gradients in deep RNN to propagate more effectively. The proposed deep RNN model was tested on a static BP dataset, and it achieved root mean square error (RMSE) of 3.90 and 2.66 mmHg for systolic BP (SBP) and diastolic BP (DBP) prediction respectively, surpassing the accuracy of traditional BP prediction models. On a multi-day BP dataset, the deep RNN achieved RMSE of 3.84, 5.25, 5.80 and 5.81 mmHg for the 1st day, 2nd day, 4th day and 6th month after the 1st day SBP prediction, and 1.80, 4.78, 5.0, 5.21 mmHg for corresponding DBP prediction, respectively, which outperforms all previous models with notable improvement. The experimental results suggest that modeling the temporal dependencies in BP dynamics significantly improves the long-term BP prediction accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge