Zhaoyang Xu
Vision Mamba in Remote Sensing: A Comprehensive Survey of Techniques, Applications and Outlook
May 01, 2025Abstract:Deep learning has profoundly transformed remote sensing, yet prevailing architectures like Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs) remain constrained by critical trade-offs: CNNs suffer from limited receptive fields, while ViTs grapple with quadratic computational complexity, hindering their scalability for high-resolution remote sensing data. State Space Models (SSMs), particularly the recently proposed Mamba architecture, have emerged as a paradigm-shifting solution, combining linear computational scaling with global context modeling. This survey presents a comprehensive review of Mamba-based methodologies in remote sensing, systematically analyzing about 120 studies to construct a holistic taxonomy of innovations and applications. Our contributions are structured across five dimensions: (i) foundational principles of vision Mamba architectures, (ii) micro-architectural advancements such as adaptive scan strategies and hybrid SSM formulations, (iii) macro-architectural integrations, including CNN-Transformer-Mamba hybrids and frequency-domain adaptations, (iv) rigorous benchmarking against state-of-the-art methods in multiple application tasks, such as object detection, semantic segmentation, change detection, etc. and (v) critical analysis of unresolved challenges with actionable future directions. By bridging the gap between SSM theory and remote sensing practice, this survey establishes Mamba as a transformative framework for remote sensing analysis. To our knowledge, this paper is the first systematic review of Mamba architectures in remote sensing. Our work provides a structured foundation for advancing research in remote sensing systems through SSM-based methods. We curate an open-source repository (https://github.com/BaoBao0926/Awesome-Mamba-in-Remote-Sensing) to foster community-driven advancements.
ASP-VMUNet: Atrous Shifted Parallel Vision Mamba U-Net for Skin Lesion Segmentation
Mar 25, 2025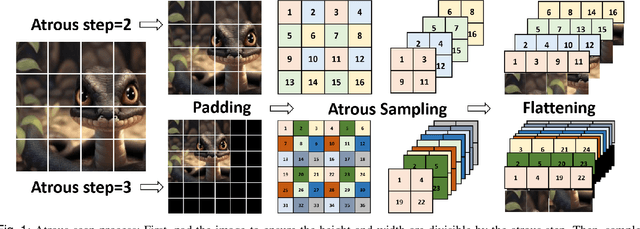
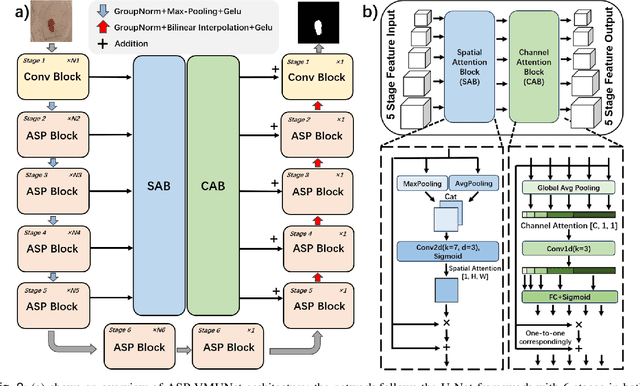
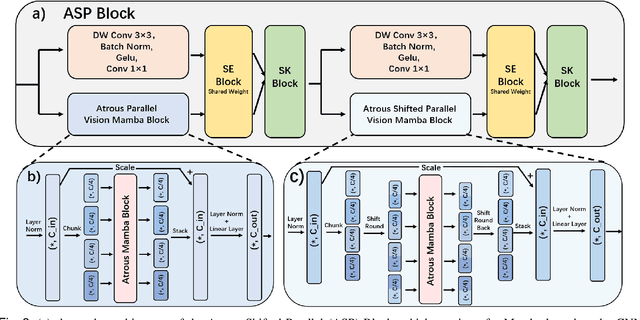
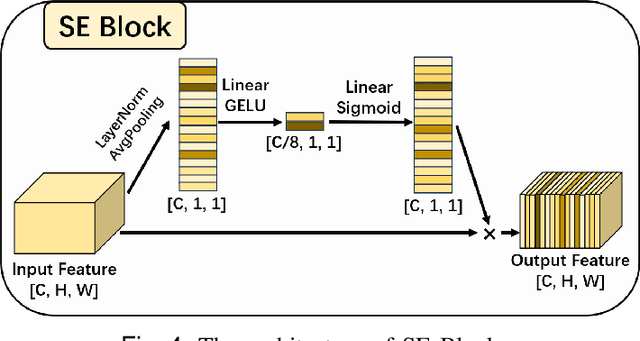
Abstract:Skin lesion segmentation is a critical challenge in computer vision, and it is essential to separate pathological features from healthy skin for diagnostics accurately. Traditional Convolutional Neural Networks (CNNs) are limited by narrow receptive fields, and Transformers face significant computational burdens. This paper presents a novel skin lesion segmentation framework, the Atrous Shifted Parallel Vision Mamba UNet (ASP-VMUNet), which integrates the efficient and scalable Mamba architecture to overcome limitations in traditional CNNs and computationally demanding Transformers. The framework introduces an atrous scan technique that minimizes background interference and expands the receptive field, enhancing Mamba's scanning capabilities. Additionally, the inclusion of a Parallel Vision Mamba (PVM) layer and a shift round operation optimizes feature segmentation and fosters rich inter-segment information exchange. A supplementary CNN branch with a Selective-Kernel (SK) Block further refines the segmentation by blending local and global contextual information. Tested on four benchmark datasets (ISIC16/17/18 and PH2), ASP-VMUNet demonstrates superior performance in skin lesion segmentation, validated by comprehensive ablation studies. This approach not only advances medical image segmentation but also highlights the benefits of hybrid architectures in medical imaging technology. Our code is available at https://github.com/BaoBao0926/ASP-VMUNet/tree/main.
Efficient Dataset Distillation via Diffusion-Driven Patch Selection for Improved Generalization
Dec 13, 2024



Abstract:Dataset distillation offers an efficient way to reduce memory and computational costs by optimizing a smaller dataset with performance comparable to the full-scale original. However, for large datasets and complex deep networks (e.g., ImageNet-1K with ResNet-101), the extensive optimization space limits performance, reducing its practicality. Recent approaches employ pre-trained diffusion models to generate informative images directly, avoiding pixel-level optimization and achieving notable results. However, these methods often face challenges due to distribution shifts between pre-trained models and target datasets, along with the need for multiple distillation steps across varying settings. To address these issues, we propose a novel framework orthogonal to existing diffusion-based distillation methods, leveraging diffusion models for selection rather than generation. Our method starts by predicting noise generated by the diffusion model based on input images and text prompts (with or without label text), then calculates the corresponding loss for each pair. With the loss differences, we identify distinctive regions of the original images. Additionally, we perform intra-class clustering and ranking on selected patches to maintain diversity constraints. This streamlined framework enables a single-step distillation process, and extensive experiments demonstrate that our approach outperforms state-of-the-art methods across various metrics.
Apple Intelligence Foundation Language Models
Jul 29, 2024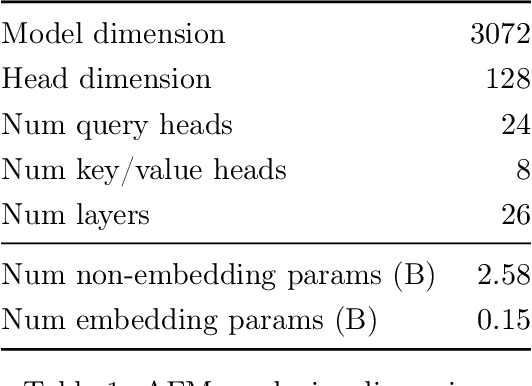
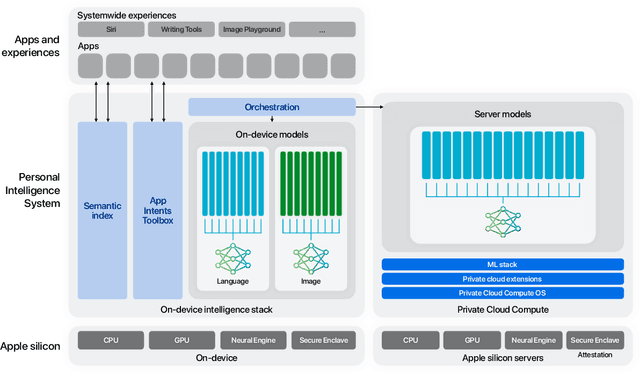


Abstract:We present foundation language models developed to power Apple Intelligence features, including a ~3 billion parameter model designed to run efficiently on devices and a large server-based language model designed for Private Cloud Compute. These models are designed to perform a wide range of tasks efficiently, accurately, and responsibly. This report describes the model architecture, the data used to train the model, the training process, how the models are optimized for inference, and the evaluation results. We highlight our focus on Responsible AI and how the principles are applied throughout the model development.
DEFN: Dual-Encoder Fourier Group Harmonics Network for Three-Dimensional Macular Hole Reconstruction with Stochastic Retinal Defect Augmentation and Dynamic Weight Composition
Nov 01, 2023Abstract:The spatial and quantitative parameters of macular holes are vital for diagnosis, surgical choices, and post-op monitoring. Macular hole diagnosis and treatment rely heavily on spatial and quantitative data, yet the scarcity of such data has impeded the progress of deep learning techniques for effective segmentation and real-time 3D reconstruction. To address this challenge, we assembled the world's largest macular hole dataset, Retinal OCTfor Macular Hole Enhancement (ROME-3914), and a Comprehensive Archive for Retinal Segmentation (CARS-30k), both expertly annotated. In addition, we developed an innovative 3D segmentation network, the Dual-Encoder FuGH Network (DEFN), which integrates three innovative modules: Fourier Group Harmonics (FuGH), Simplified 3D Spatial Attention (S3DSA) and Harmonic Squeeze-and-Excitation Module (HSE). These three modules synergistically filter noise, reduce computational complexity, emphasize detailed features, and enhance the network's representation ability. We also proposed a novel data augmentation method, Stochastic Retinal Defect Injection (SRDI), and a network optimization strategy DynamicWeightCompose (DWC), to further improve the performance of DEFN. Compared with 13 baselines, our DEFN shows the best performance. We also offer precise 3D retinal reconstruction and quantitative metrics, bringing revolutionary diagnostic and therapeutic decision-making tools for ophthalmologists, and is expected to completely reshape the diagnosis and treatment patterns of difficult-to-treat macular degeneration. The source code is publicly available at: https://github.com/IIPL-HangzhouDianUniversity/DEFN-Pytorch.
Change Detection Methods for Remote Sensing in the Last Decade: A Comprehensive Review
May 09, 2023



Abstract:Change detection is an essential and widely utilized task in remote sensing that aims to detect and analyze changes occurring in the same geographical area over time, which has broad applications in urban development, agricultural surveys, and land cover monitoring. Detecting changes in remote sensing images is a complex challenge due to various factors, including variations in image quality, noise, registration errors, illumination changes, complex landscapes, and spatial heterogeneity. In recent years, deep learning has emerged as a powerful tool for feature extraction and addressing these challenges. Its versatility has resulted in its widespread adoption for numerous image-processing tasks. This paper presents a comprehensive survey of significant advancements in change detection for remote sensing images over the past decade. We first introduce some preliminary knowledge for the change detection task, such as problem definition, datasets, evaluation metrics, and transformer basics, as well as provide a detailed taxonomy of existing algorithms from three different perspectives: algorithm granularity, supervision modes, and learning frameworks in the methodology section. This survey enables readers to gain systematic knowledge of change detection tasks from various angles. We then summarize the state-of-the-art performance on several dominant change detection datasets, providing insights into the strengths and limitations of existing algorithms. Based on our survey, some future research directions for change detection in remote sensing are well identified. This survey paper will shed some light on the community and inspire further research efforts in the change detection task.
Interpretable Survival Prediction for Colorectal Cancer using Deep Learning
Nov 17, 2020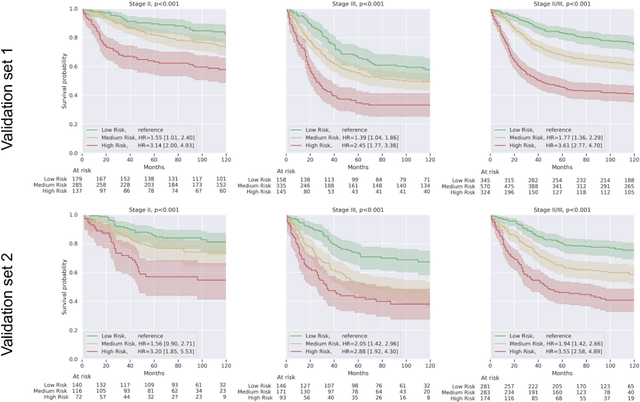
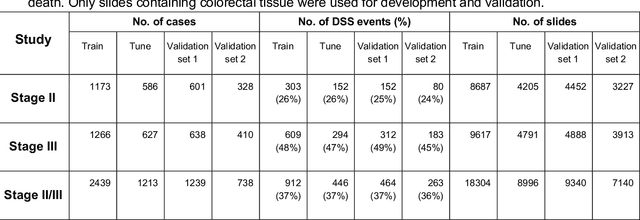
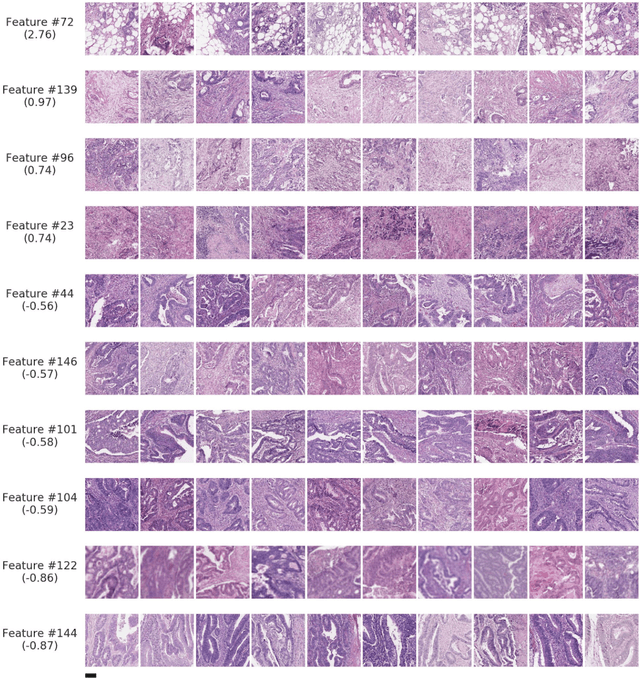
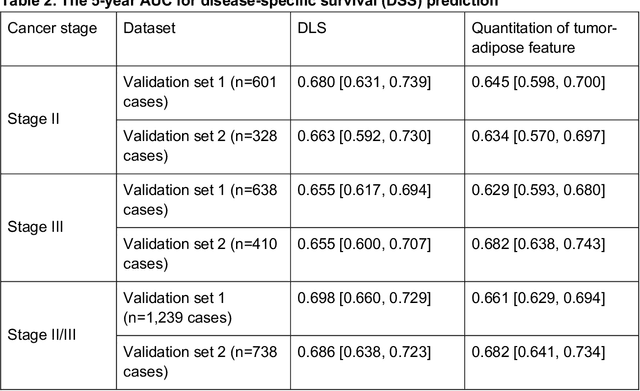
Abstract:Deriving interpretable prognostic features from deep-learning-based prognostic histopathology models remains a challenge. In this study, we developed a deep learning system (DLS) for predicting disease specific survival for stage II and III colorectal cancer using 3,652 cases (27,300 slides). When evaluated on two validation datasets containing 1,239 cases (9,340 slides) and 738 cases (7,140 slides) respectively, the DLS achieved a 5-year disease-specific survival AUC of 0.70 (95%CI 0.66-0.73) and 0.69 (95%CI 0.64-0.72), and added significant predictive value to a set of 9 clinicopathologic features. To interpret the DLS, we explored the ability of different human-interpretable features to explain the variance in DLS scores. We observed that clinicopathologic features such as T-category, N-category, and grade explained a small fraction of the variance in DLS scores (R2=18% in both validation sets). Next, we generated human-interpretable histologic features by clustering embeddings from a deep-learning based image-similarity model and showed that they explain the majority of the variance (R2 of 73% to 80%). Furthermore, the clustering-derived feature most strongly associated with high DLS scores was also highly prognostic in isolation. With a distinct visual appearance (poorly differentiated tumor cell clusters adjacent to adipose tissue), this feature was identified by annotators with 87.0-95.5% accuracy. Our approach can be used to explain predictions from a prognostic deep learning model and uncover potentially-novel prognostic features that can be reliably identified by people for future validation studies.
Deep learning-based survival prediction for multiple cancer types using histopathology images
Dec 16, 2019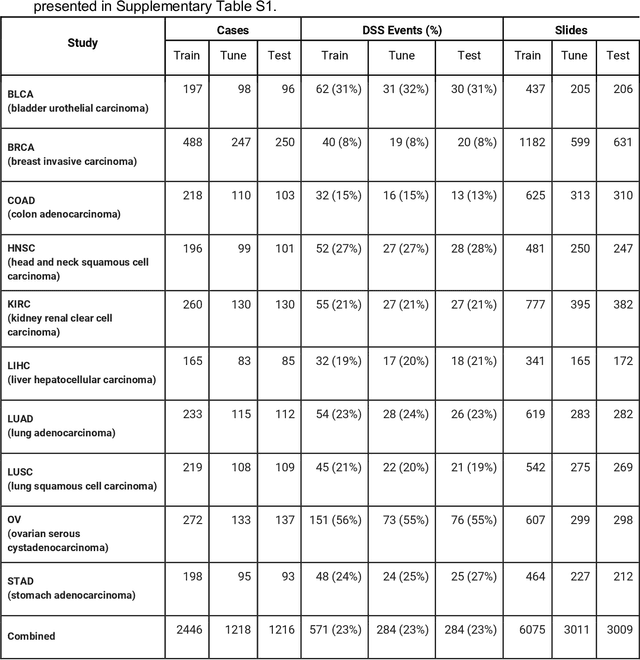
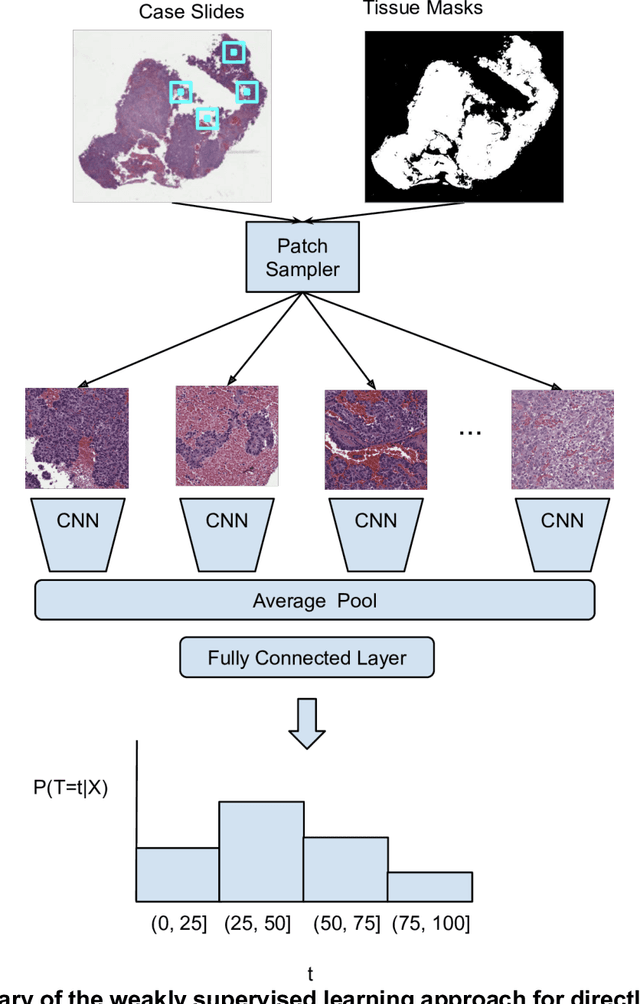
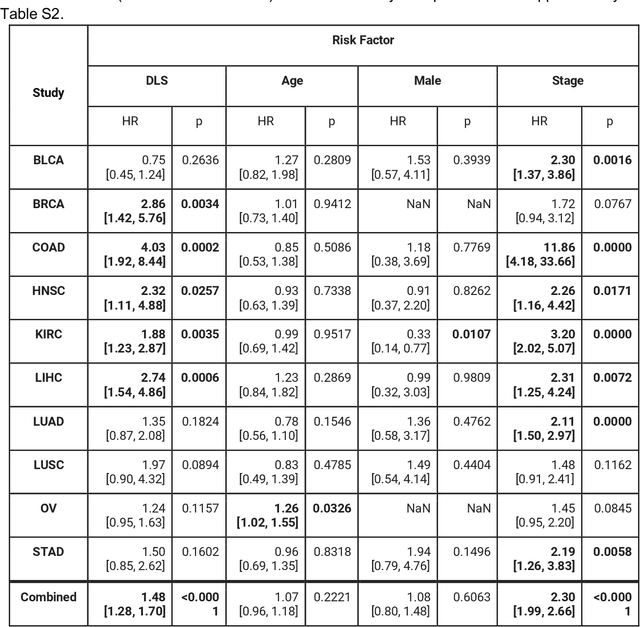
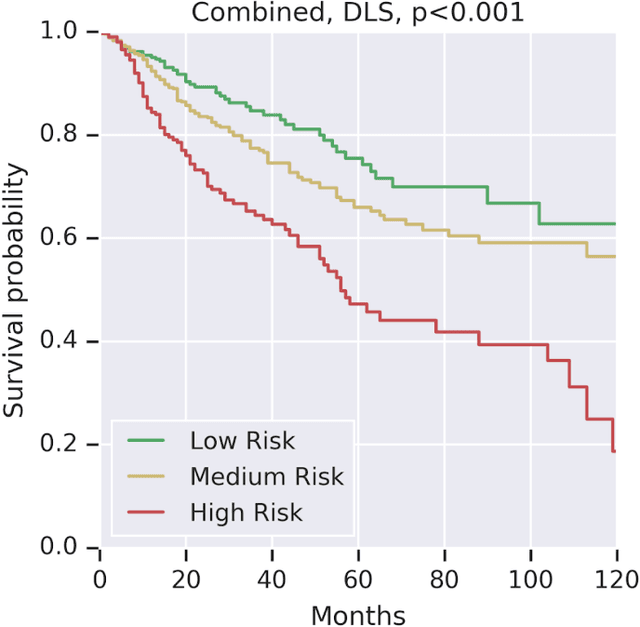
Abstract:Prognostic information at diagnosis has important implications for cancer treatment and monitoring. Although cancer staging, histopathological assessment, molecular features, and clinical variables can provide useful prognostic insights, improving risk stratification remains an active research area. We developed a deep learning system (DLS) to predict disease specific survival across 10 cancer types from The Cancer Genome Atlas (TCGA). We used a weakly-supervised approach without pixel-level annotations, and tested three different survival loss functions. The DLS was developed using 9,086 slides from 3,664 cases and evaluated using 3,009 slides from 1,216 cases. In multivariable Cox regression analysis of the combined cohort including all 10 cancers, the DLS was significantly associated with disease specific survival (hazard ratio of 1.58, 95% CI 1.28-1.70, p<0.0001) after adjusting for cancer type, stage, age, and sex. In a per-cancer adjusted subanalysis, the DLS remained a significant predictor of survival in 5 of 10 cancer types. Compared to a baseline model including stage, age, and sex, the c-index of the model demonstrated an absolute 3.7% improvement (95% CI 1.0-6.5) in the combined cohort. Additionally, our models stratified patients within individual cancer stages, particularly stage II (p=0.025) and stage III (p<0.001). By developing and evaluating prognostic models across multiple cancer types, this work represents one of the most comprehensive studies exploring the direct prediction of clinical outcomes using deep learning and histopathology images. Our analysis demonstrates the potential for this approach to provide prognostic information in multiple cancer types, and even within specific pathologic stages. However, given the relatively small number of clinical events, we observed wide confidence intervals, suggesting that future work will benefit from larger datasets.
US-net for robust and efficient nuclei instance segmentation
Jan 31, 2019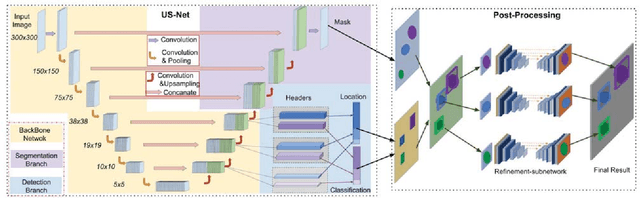
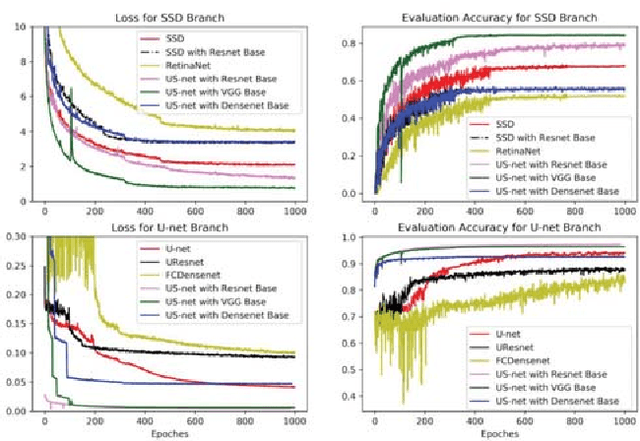
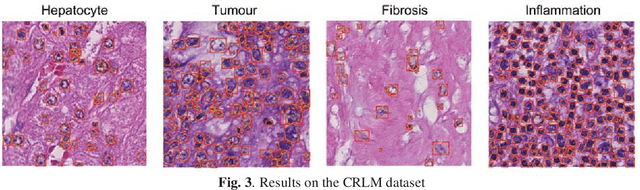
Abstract:We present a novel neural network architecture, US-Net, for robust nuclei instance segmentation in histopathology images. The proposed framework integrates the nuclei detection and segmentation networks by sharing their outputs through the same foundation network, and thus enhancing the performance of both. The detection network takes into account the high-level semantic cues with contextual information, while the segmentation network focuses more on the low-level details like the edges. Extensive experiments reveal that our proposed framework can strengthen the performance of both branch networks in an integrated architecture and outperforms most of the state-of-the-art nuclei detection and segmentation networks.
GAN-based Virtual Re-Staining: A Promising Solution for Whole Slide Image Analysis
Jan 13, 2019



Abstract:Histopathological cancer diagnosis is based on visual examination of stained tissue slides. Hematoxylin and eosin (H\&E) is a standard stain routinely employed worldwide. It is easy to acquire and cost effective, but cells and tissue components show low-contrast with varying tones of dark blue and pink, which makes difficult visual assessments, digital image analysis, and quantifications. These limitations can be overcome by IHC staining of target proteins of the tissue slide. IHC provides a selective, high-contrast imaging of cells and tissue components, but their use is largely limited by a significantly more complex laboratory processing and high cost. We proposed a conditional CycleGAN (cCGAN) network to transform the H\&E stained images into IHC stained images, facilitating virtual IHC staining on the same slide. This data-driven method requires only a limited amount of labelled data but will generate pixel level segmentation results. The proposed cCGAN model improves the original network \cite{zhu_unpaired_2017} by adding category conditions and introducing two structural loss functions, which realize a multi-subdomain translation and improve the translation accuracy as well. % need to give reasons here. Experiments demonstrate that the proposed model outperforms the original method in unpaired image translation with multi-subdomains. We also explore the potential of unpaired images to image translation method applied on other histology images related tasks with different staining techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge