Xingru Huang
BusterX: MLLM-Powered AI-Generated Video Forgery Detection and Explanation
May 19, 2025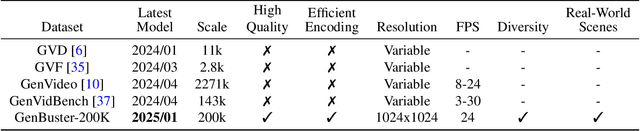
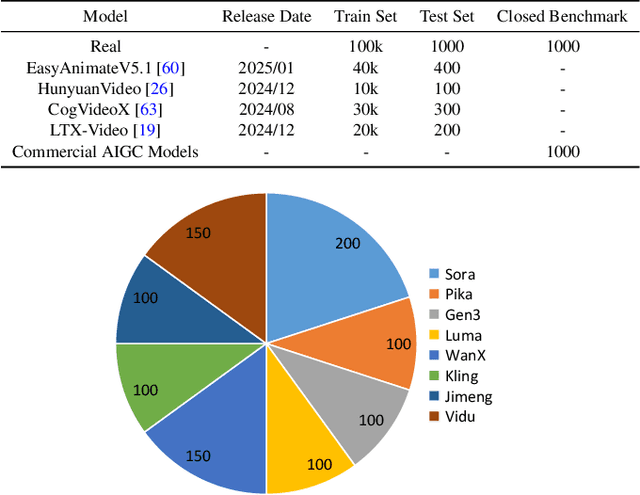
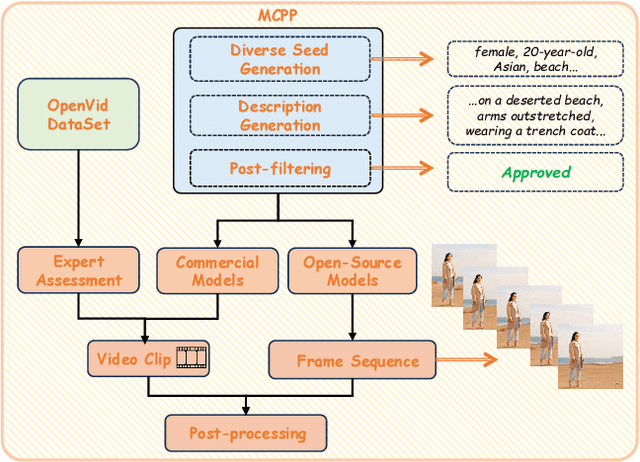
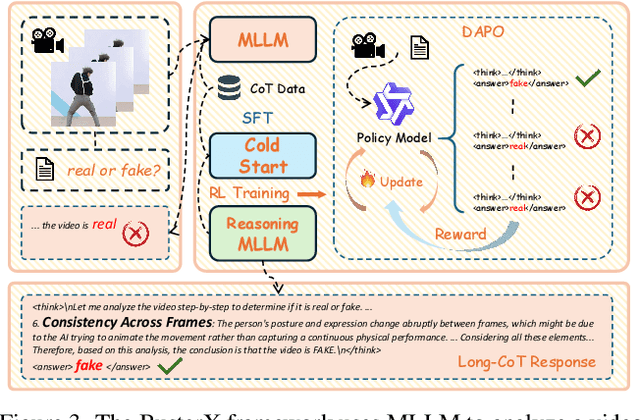
Abstract:Advances in AI generative models facilitate super-realistic video synthesis, amplifying misinformation risks via social media and eroding trust in digital content. Several research works have explored new deepfake detection methods on AI-generated images to alleviate these risks. However, with the fast development of video generation models, such as Sora and WanX, there is currently a lack of large-scale, high-quality AI-generated video datasets for forgery detection. In addition, existing detection approaches predominantly treat the task as binary classification, lacking explainability in model decision-making and failing to provide actionable insights or guidance for the public. To address these challenges, we propose \textbf{GenBuster-200K}, a large-scale AI-generated video dataset featuring 200K high-resolution video clips, diverse latest generative techniques, and real-world scenes. We further introduce \textbf{BusterX}, a novel AI-generated video detection and explanation framework leveraging multimodal large language model (MLLM) and reinforcement learning for authenticity determination and explainable rationale. To our knowledge, GenBuster-200K is the {\it \textbf{first}} large-scale, high-quality AI-generated video dataset that incorporates the latest generative techniques for real-world scenarios. BusterX is the {\it \textbf{first}} framework to integrate MLLM with reinforcement learning for explainable AI-generated video detection. Extensive comparisons with state-of-the-art methods and ablation studies validate the effectiveness and generalizability of BusterX. The code, models, and datasets will be released.
DEFN: Dual-Encoder Fourier Group Harmonics Network for Three-Dimensional Macular Hole Reconstruction with Stochastic Retinal Defect Augmentation and Dynamic Weight Composition
Nov 01, 2023Abstract:The spatial and quantitative parameters of macular holes are vital for diagnosis, surgical choices, and post-op monitoring. Macular hole diagnosis and treatment rely heavily on spatial and quantitative data, yet the scarcity of such data has impeded the progress of deep learning techniques for effective segmentation and real-time 3D reconstruction. To address this challenge, we assembled the world's largest macular hole dataset, Retinal OCTfor Macular Hole Enhancement (ROME-3914), and a Comprehensive Archive for Retinal Segmentation (CARS-30k), both expertly annotated. In addition, we developed an innovative 3D segmentation network, the Dual-Encoder FuGH Network (DEFN), which integrates three innovative modules: Fourier Group Harmonics (FuGH), Simplified 3D Spatial Attention (S3DSA) and Harmonic Squeeze-and-Excitation Module (HSE). These three modules synergistically filter noise, reduce computational complexity, emphasize detailed features, and enhance the network's representation ability. We also proposed a novel data augmentation method, Stochastic Retinal Defect Injection (SRDI), and a network optimization strategy DynamicWeightCompose (DWC), to further improve the performance of DEFN. Compared with 13 baselines, our DEFN shows the best performance. We also offer precise 3D retinal reconstruction and quantitative metrics, bringing revolutionary diagnostic and therapeutic decision-making tools for ophthalmologists, and is expected to completely reshape the diagnosis and treatment patterns of difficult-to-treat macular degeneration. The source code is publicly available at: https://github.com/IIPL-HangzhouDianUniversity/DEFN-Pytorch.
Structure-aware scale-adaptive networks for cancer segmentation in whole-slide images
Sep 26, 2021



Abstract:Cancer segmentation in whole-slide images is a fundamental step for viable tumour burden estimation, which is of great value for cancer assessment. However, factors like vague boundaries or small regions dissociated from viable tumour areas make it a challenging task. Considering the usefulness of multi-scale features in various vision-related tasks, we present a structure-aware scale-adaptive feature selection method for efficient and accurate cancer segmentation. Based on a segmentation network with a popular encoder-decoder architecture, a scale-adaptive module is proposed for selecting more robust features to represent the vague, non-rigid boundaries. Furthermore, a structural similarity metric is proposed for better tissue structure awareness to deal with small region segmentation. In addition, advanced designs including several attention mechanisms and the selective-kernel convolutions are applied to the baseline network for comparative study purposes. Extensive experimental results show that the proposed structure-aware scale-adaptive networks achieve outstanding performance on liver cancer segmentation when compared to top ten submitted results in the challenge of PAIP 2019. Further evaluation on colorectal cancer segmentation shows that the scale-adaptive module improves the baseline network or outperforms the other excellent designs of attention mechanisms when considering the tradeoff between efficiency and accuracy.
Magnification-independent Histopathological Image Classification with Similarity-based Multi-scale Embeddings
Jul 02, 2021



Abstract:The classification of histopathological images is of great value in both cancer diagnosis and pathological studies. However, multiple reasons, such as variations caused by magnification factors and class imbalance, make it a challenging task where conventional methods that learn from image-label datasets perform unsatisfactorily in many cases. We observe that tumours of the same class often share common morphological patterns. To exploit this fact, we propose an approach that learns similarity-based multi-scale embeddings (SMSE) for magnification-independent histopathological image classification. In particular, a pair loss and a triplet loss are leveraged to learn similarity-based embeddings from image pairs or image triplets. The learned embeddings provide accurate measurements of similarities between images, which are regarded as a more effective form of representation for histopathological morphology than normal image features. Furthermore, in order to ensure the generated models are magnification-independent, images acquired at different magnification factors are simultaneously fed to networks during training for learning multi-scale embeddings. In addition to the SMSE, to eliminate the impact of class imbalance, instead of using the hard sample mining strategy that intuitively discards some easy samples, we introduce a new reinforced focal loss to simultaneously punish hard misclassified samples while suppressing easy well-classified samples. Experimental results show that the SMSE improves the performance for histopathological image classification tasks for both breast and liver cancers by a large margin compared to previous methods. In particular, the SMSE achieves the best performance on the BreakHis benchmark with an improvement ranging from 5% to 18% compared to previous methods using traditional features.
SRPN: similarity-based region proposal networks for nuclei and cells detection in histology images
Jun 25, 2021



Abstract:The detection of nuclei and cells in histology images is of great value in both clinical practice and pathological studies. However, multiple reasons such as morphological variations of nuclei or cells make it a challenging task where conventional object detection methods cannot obtain satisfactory performance in many cases. A detection task consists of two sub-tasks, classification and localization. Under the condition of dense object detection, classification is a key to boost the detection performance. Considering this, we propose similarity based region proposal networks (SRPN) for nuclei and cells detection in histology images. In particular, a customized convolution layer termed as embedding layer is designed for network building. The embedding layer is added into the region proposal networks, enabling the networks to learn discriminative features based on similarity learning. Features obtained by similarity learning can significantly boost the classification performance compared to conventional methods. SRPN can be easily integrated into standard convolutional neural networks architectures such as the Faster R-CNN and RetinaNet. We test the proposed approach on tasks of multi-organ nuclei detection and signet ring cells detection in histological images. Experimental results show that networks applying similarity learning achieved superior performance on both tasks when compared to their counterparts. In particular, the proposed SRPN achieve state-of-the-art performance on the MoNuSeg benchmark for nuclei segmentation and detection while compared to previous methods, and on the signet ring cell detection benchmark when compared with baselines. The sourcecode is publicly available at: https://github.com/sigma10010/nuclei_cells_det.
* Accepted by Medical Image Analysis for publication
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge