Yen-Wei Chen
A Text-Image Fusion Method with Data Augmentation Capabilities for Referring Medical Image Segmentation
Oct 14, 2025



Abstract:Deep learning relies heavily on data augmentation to mitigate limited data, especially in medical imaging. Recent multimodal learning integrates text and images for segmentation, known as referring or text-guided image segmentation. However, common augmentations like rotation and flipping disrupt spatial alignment between image and text, weakening performance. To address this, we propose an early fusion framework that combines text and visual features before augmentation, preserving spatial consistency. We also design a lightweight generator that projects text embeddings into visual space, bridging semantic gaps. Visualization of generated pseudo-images shows accurate region localization. Our method is evaluated on three medical imaging tasks and four segmentation frameworks, achieving state-of-the-art results. Code is publicly available on GitHub: https://github.com/11yxk/MedSeg_EarlyFusion.
EPIC: Efficient Prompt Interaction for Text-Image Classification
Jul 10, 2025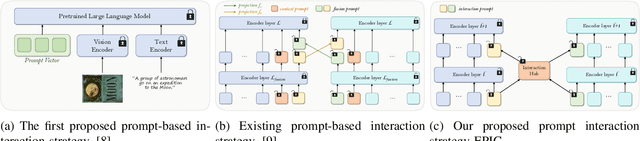
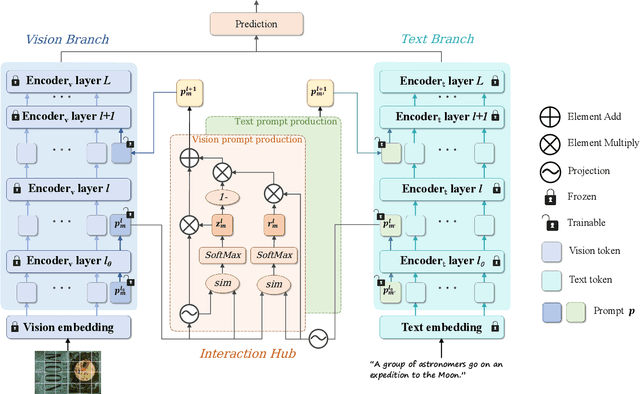
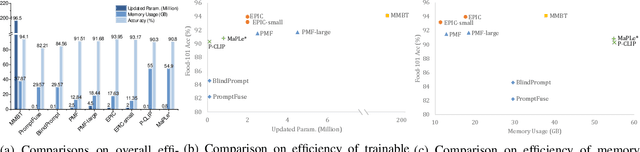
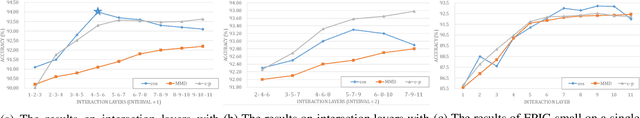
Abstract:In recent years, large-scale pre-trained multimodal models (LMMs) generally emerge to integrate the vision and language modalities, achieving considerable success in multimodal tasks, such as text-image classification. The growing size of LMMs, however, results in a significant computational cost for fine-tuning these models for downstream tasks. Hence, prompt-based interaction strategy is studied to align modalities more efficiently. In this context, we propose a novel efficient prompt-based multimodal interaction strategy, namely Efficient Prompt Interaction for text-image Classification (EPIC). Specifically, we utilize temporal prompts on intermediate layers, and integrate different modalities with similarity-based prompt interaction, to leverage sufficient information exchange between modalities. Utilizing this approach, our method achieves reduced computational resource consumption and fewer trainable parameters (about 1\% of the foundation model) compared to other fine-tuning strategies. Furthermore, it demonstrates superior performance on the UPMC-Food101 and SNLI-VE datasets, while achieving comparable performance on the MM-IMDB dataset.
SemSim: Revisiting Weak-to-Strong Consistency from a Semantic Similarity Perspective for Semi-supervised Medical Image Segmentation
Oct 17, 2024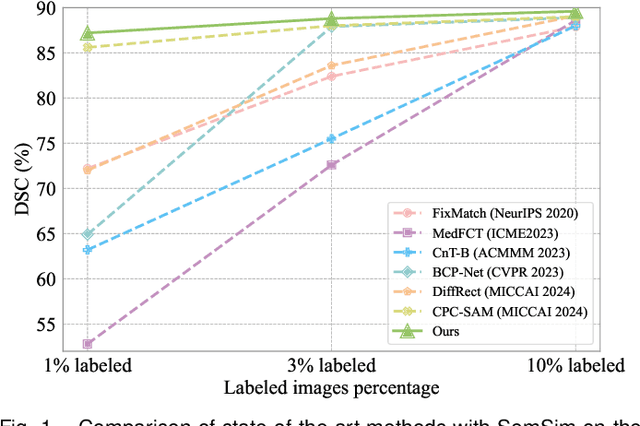
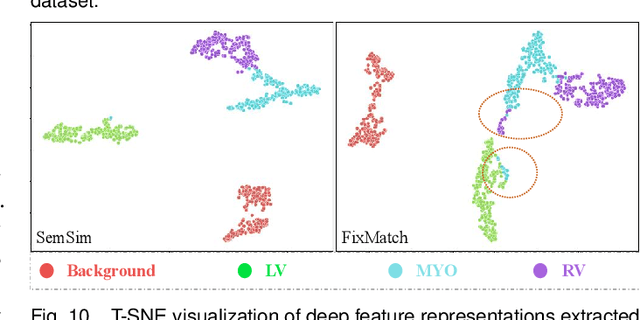
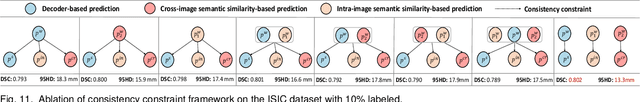
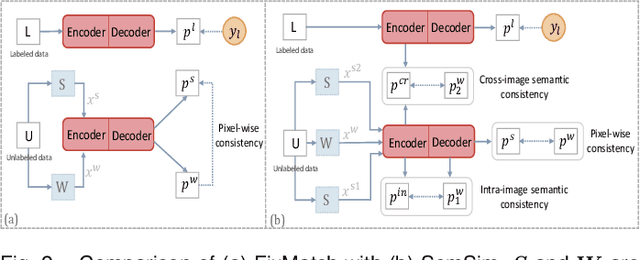
Abstract:Semi-supervised learning (SSL) for medical image segmentation is a challenging yet highly practical task, which reduces reliance on large-scale labeled dataset by leveraging unlabeled samples. Among SSL techniques, the weak-to-strong consistency framework, popularized by FixMatch, has emerged as a state-of-the-art method in classification tasks. Notably, such a simple pipeline has also shown competitive performance in medical image segmentation. However, two key limitations still persist, impeding its efficient adaptation: (1) the neglect of contextual dependencies results in inconsistent predictions for similar semantic features, leading to incomplete object segmentation; (2) the lack of exploitation of semantic similarity between labeled and unlabeled data induces considerable class-distribution discrepancy. To address these limitations, we propose a novel semi-supervised framework based on FixMatch, named SemSim, powered by two appealing designs from semantic similarity perspective: (1) rectifying pixel-wise prediction by reasoning about the intra-image pair-wise affinity map, thus integrating contextual dependencies explicitly into the final prediction; (2) bridging labeled and unlabeled data via a feature querying mechanism for compact class representation learning, which fully considers cross-image anatomical similarities. As the reliable semantic similarity extraction depends on robust features, we further introduce an effective spatial-aware fusion module (SFM) to explore distinctive information from multiple scales. Extensive experiments show that SemSim yields consistent improvements over the state-of-the-art methods across three public segmentation benchmarks.
Deep Self-cleansing for Medical Image Segmentation with Noisy Labels
Sep 08, 2024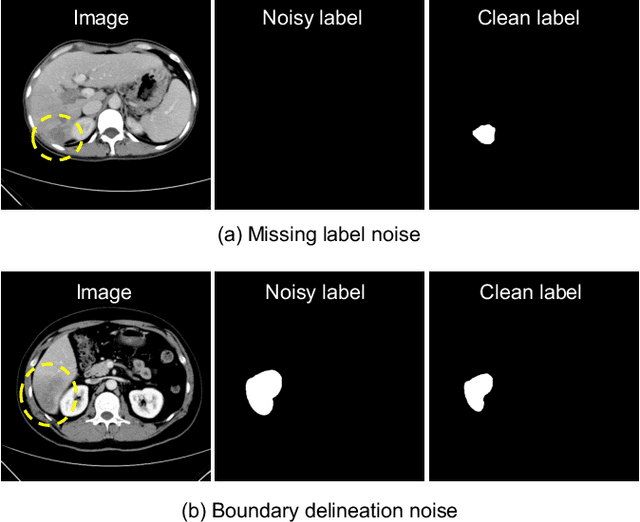
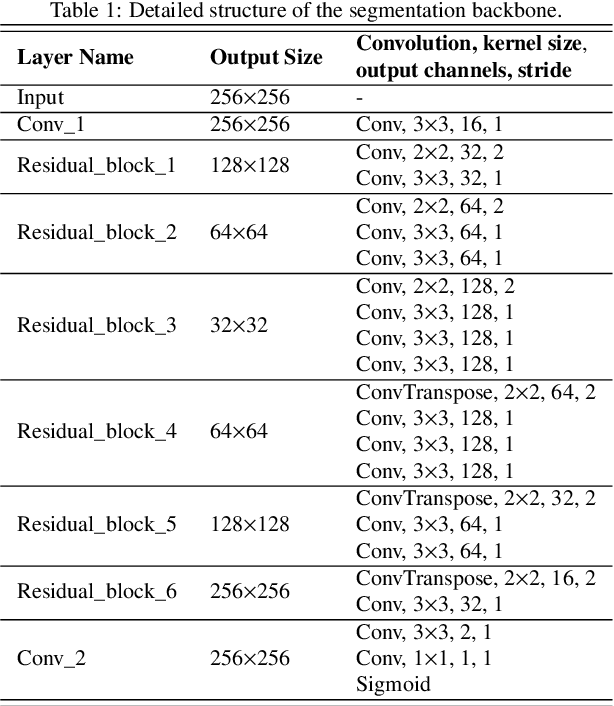
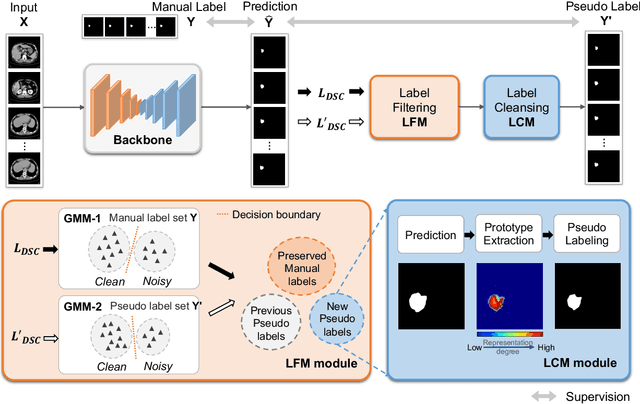
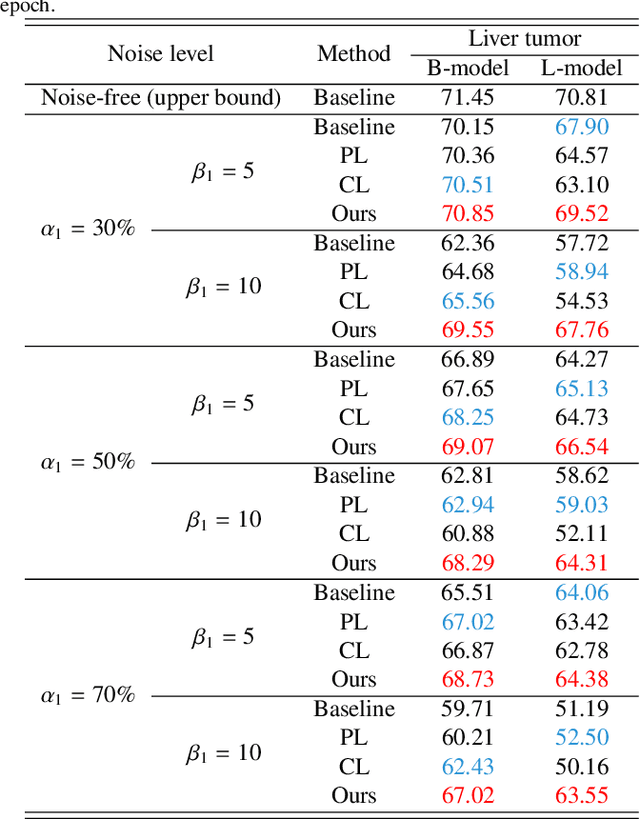
Abstract:Medical image segmentation is crucial in the field of medical imaging, aiding in disease diagnosis and surgical planning. Most established segmentation methods rely on supervised deep learning, in which clean and precise labels are essential for supervision and significantly impact the performance of models. However, manually delineated labels often contain noise, such as missing labels and inaccurate boundary delineation, which can hinder networks from correctly modeling target characteristics. In this paper, we propose a deep self-cleansing segmentation framework that can preserve clean labels while cleansing noisy ones in the training phase. To achieve this, we devise a gaussian mixture model-based label filtering module that distinguishes noisy labels from clean labels. Additionally, we develop a label cleansing module to generate pseudo low-noise labels for identified noisy samples. The preserved clean labels and pseudo-labels are then used jointly to supervise the network. Validated on a clinical liver tumor dataset and a public cardiac diagnosis dataset, our method can effectively suppress the interference from noisy labels and achieve prominent segmentation performance.
LSMS: Language-guided Scale-aware MedSegmentor for Medical Image Referring Segmentation
Sep 02, 2024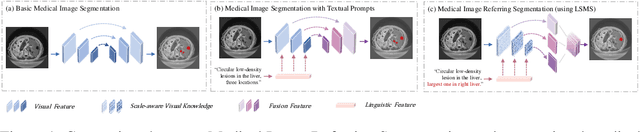
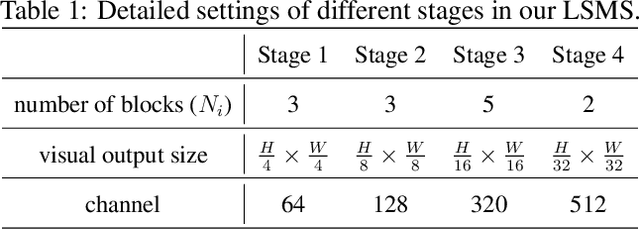
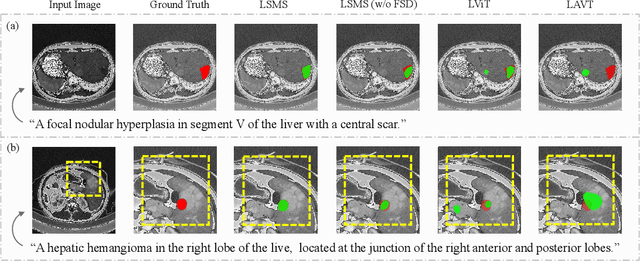
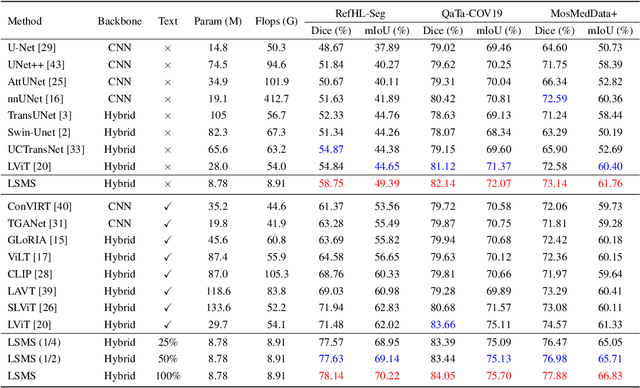
Abstract:Conventional medical image segmentation methods have been found inadequate in facilitating physicians with the identification of specific lesions for diagnosis and treatment. Given the utility of text as an instructional format, we introduce a novel task termed Medical Image Referring Segmentation (MIRS), which requires segmenting specified lesions in images based on the given language expressions. Due to the varying object scales in medical images, MIRS demands robust vision-language modeling and comprehensive multi-scale interaction for precise localization and segmentation under linguistic guidance. However, existing medical image segmentation methods fall short in meeting these demands, resulting in insufficient segmentation accuracy. In response, we propose an approach named Language-guided Scale-aware MedSegmentor (LSMS), incorporating two appealing designs: (1)~a Scale-aware Vision-Language Attention module that leverages diverse convolutional kernels to acquire rich visual knowledge and interact closely with linguistic features, thereby enhancing lesion localization capability; (2)~a Full-Scale Decoder that globally models multi-modal features across various scales, capturing complementary information between scales to accurately outline lesion boundaries. Addressing the lack of suitable datasets for MIRS, we constructed a vision-language medical dataset called Reference Hepatic Lesion Segmentation (RefHL-Seg). This dataset comprises 2,283 abdominal CT slices from 231 cases, with corresponding textual annotations and segmentation masks for various liver lesions in images. We validated the performance of LSMS for MIRS and conventional medical image segmentation tasks across various datasets. Our LSMS consistently outperforms on all datasets with lower computational costs. The code and datasets will be released.
Multitask and Multimodal Neural Tuning for Large Models
Aug 06, 2024



Abstract:In recent years, large-scale multimodal models have demonstrated impressive capabilities across various domains. However, enabling these models to effectively perform multiple multimodal tasks simultaneously remains a significant challenge. To address this, we introduce a novel tuning method called neural tuning, designed to handle diverse multimodal tasks concurrently, including reasoning segmentation, referring segmentation, image captioning, and text-to-image generation. Neural tuning emulates sparse distributed representation in human brain, where only specific subsets of neurons are activated for each task. Additionally, we present a new benchmark, MMUD, where each sample is annotated with multiple task labels. By applying neural tuning to pretrained large models on the MMUD benchmark, we achieve simultaneous task handling in a streamlined and efficient manner. All models, code, and datasets will be publicly available after publication, facilitating further research and development in this field.
HSVLT: Hierarchical Scale-Aware Vision-Language Transformer for Multi-Label Image Classification
Jul 23, 2024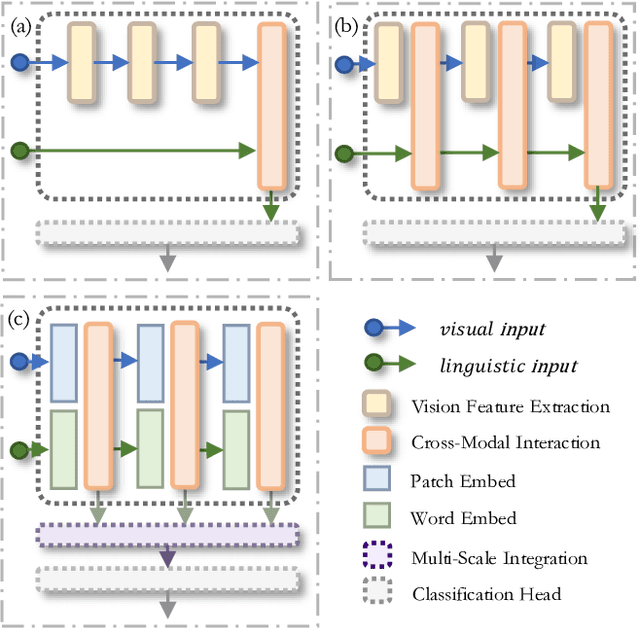
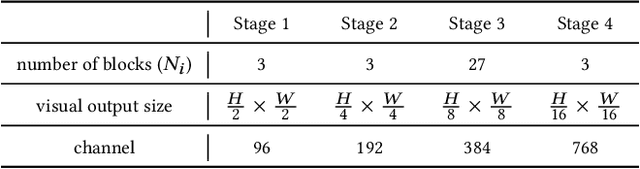
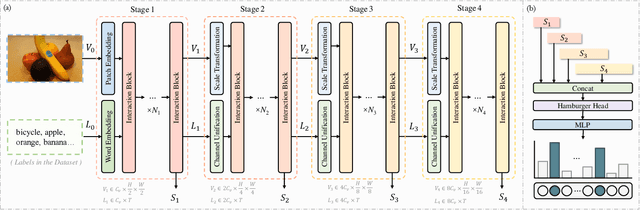
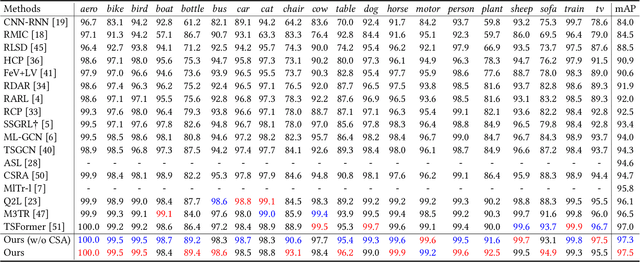
Abstract:The task of multi-label image classification involves recognizing multiple objects within a single image. Considering both valuable semantic information contained in the labels and essential visual features presented in the image, tight visual-linguistic interactions play a vital role in improving classification performance. Moreover, given the potential variance in object size and appearance within a single image, attention to features of different scales can help to discover possible objects in the image. Recently, Transformer-based methods have achieved great success in multi-label image classification by leveraging the advantage of modeling long-range dependencies, but they have several limitations. Firstly, existing methods treat visual feature extraction and cross-modal fusion as separate steps, resulting in insufficient visual-linguistic alignment in the joint semantic space. Additionally, they only extract visual features and perform cross-modal fusion at a single scale, neglecting objects with different characteristics. To address these issues, we propose a Hierarchical Scale-Aware Vision-Language Transformer (HSVLT) with two appealing designs: (1)~A hierarchical multi-scale architecture that involves a Cross-Scale Aggregation module, which leverages joint multi-modal features extracted from multiple scales to recognize objects of varying sizes and appearances in images. (2)~Interactive Visual-Linguistic Attention, a novel attention mechanism module that tightly integrates cross-modal interaction, enabling the joint updating of visual, linguistic and multi-modal features. We have evaluated our method on three benchmark datasets. The experimental results demonstrate that HSVLT surpasses state-of-the-art methods with lower computational cost.
* 10 pages, 6 figures
SOFIM: Stochastic Optimization Using Regularized Fisher Information Matrix
Mar 05, 2024



Abstract:This paper introduces a new stochastic optimization method based on the regularized Fisher information matrix (FIM), named SOFIM, which can efficiently utilize the FIM to approximate the Hessian matrix for finding Newton's gradient update in large-scale stochastic optimization of machine learning models. It can be viewed as a variant of natural gradient descent (NGD), where the challenge of storing and calculating the full FIM is addressed through making use of the regularized FIM and directly finding the gradient update direction via Sherman-Morrison matrix inversion. Additionally, like the popular Adam method, SOFIM uses the first moment of the gradient to address the issue of non-stationary objectives across mini-batches due to heterogeneous data. The utilization of the regularized FIM and Sherman-Morrison matrix inversion leads to the improved convergence rate with the same space and time complexities as stochastic gradient descent (SGD) with momentum. The extensive experiments on training deep learning models on several benchmark image classification datasets demonstrate that the proposed SOFIM outperforms SGD with momentum and several state-of-the-art Newton optimization methods, such as Nystrom-SGD, L-BFGS, and AdaHessian, in term of the convergence speed for achieving the pre-specified objectives of training and test losses as well as test accuracy.
Memory-Inspired Temporal Prompt Interaction for Text-Image Classification
Jan 26, 2024Abstract:In recent years, large-scale pre-trained multimodal models (LMM) generally emerge to integrate the vision and language modalities, achieving considerable success in various natural language processing and computer vision tasks. The growing size of LMMs, however, results in a significant computational cost for fine-tuning these models for downstream tasks. Hence, prompt-based interaction strategy is studied to align modalities more efficiently. In this contex, we propose a novel prompt-based multimodal interaction strategy inspired by human memory strategy, namely Memory-Inspired Temporal Prompt Interaction (MITP). Our proposed method involves in two stages as in human memory strategy: the acquiring stage, and the consolidation and activation stage. We utilize temporal prompts on intermediate layers to imitate the acquiring stage, leverage similarity-based prompt interaction to imitate memory consolidation, and employ prompt generation strategy to imitate memory activation. The main strength of our paper is that we interact the prompt vectors on intermediate layers to leverage sufficient information exchange between modalities, with compressed trainable parameters and memory usage. We achieve competitive results on several datasets with relatively small memory usage and 2.0M of trainable parameters (about 1% of the pre-trained foundation model).
M2ORT: Many-To-One Regression Transformer for Spatial Transcriptomics Prediction from Histopathology Images
Jan 24, 2024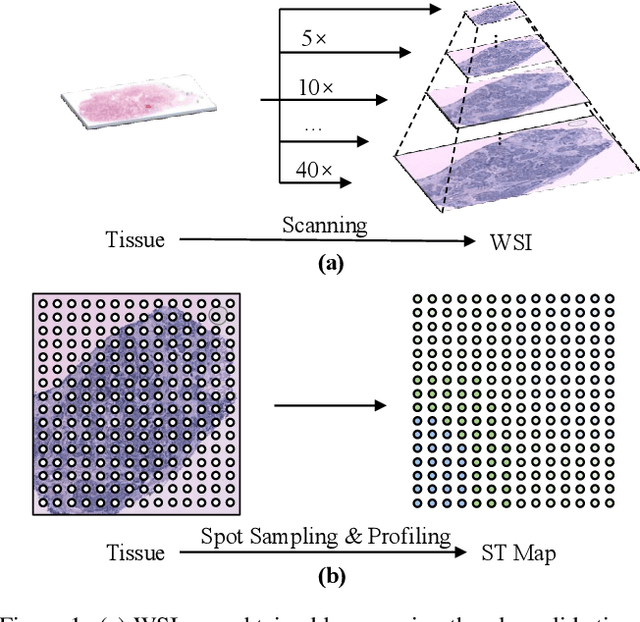
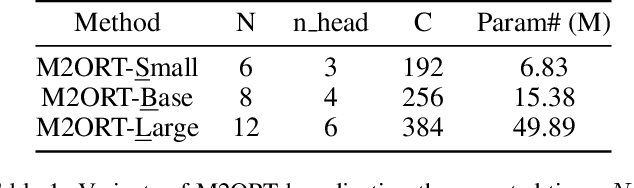
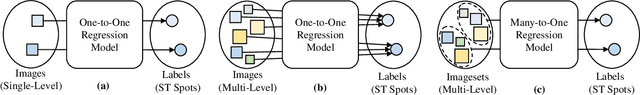
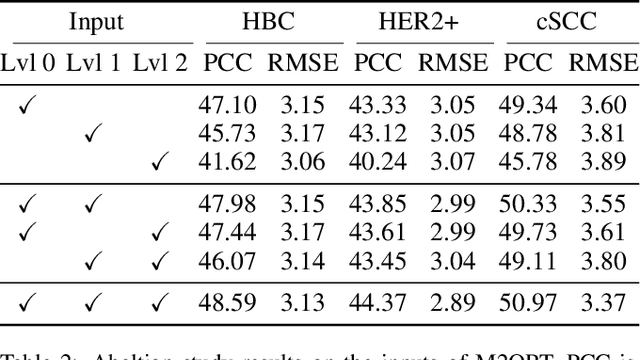
Abstract:The advancement of Spatial Transcriptomics (ST) has facilitated the spatially-aware profiling of gene expressions based on histopathology images. Although ST data offers valuable insights into the micro-environment of tumors, its acquisition cost remains expensive. Therefore, directly predicting the ST expressions from digital pathology images is desired. Current methods usually adopt existing regression backbones for this task, which ignore the inherent multi-scale hierarchical data structure of digital pathology images. To address this limit, we propose M2ORT, a many-to-one regression Transformer that can accommodate the hierarchical structure of the pathology images through a decoupled multi-scale feature extractor. Different from traditional models that are trained with one-to-one image-label pairs, M2ORT accepts multiple pathology images of different magnifications at a time to jointly predict the gene expressions at their corresponding common ST spot, aiming at learning a many-to-one relationship through training. We have tested M2ORT on three public ST datasets and the experimental results show that M2ORT can achieve state-of-the-art performance with fewer parameters and floating-point operations (FLOPs). The code is available at: https://github.com/Dootmaan/M2ORT/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge