Hongjie Hu
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
Rethinking Multiple Instance Learning for Whole Slide Image Classification: A Bag-Level Classifier is a Good Instance-Level Teacher
Dec 02, 2023Abstract:Multiple Instance Learning (MIL) has demonstrated promise in Whole Slide Image (WSI) classification. However, a major challenge persists due to the high computational cost associated with processing these gigapixel images. Existing methods generally adopt a two-stage approach, comprising a non-learnable feature embedding stage and a classifier training stage. Though it can greatly reduce the memory consumption by using a fixed feature embedder pre-trained on other domains, such scheme also results in a disparity between the two stages, leading to suboptimal classification accuracy. To address this issue, we propose that a bag-level classifier can be a good instance-level teacher. Based on this idea, we design Iteratively Coupled Multiple Instance Learning (ICMIL) to couple the embedder and the bag classifier at a low cost. ICMIL initially fix the patch embedder to train the bag classifier, followed by fixing the bag classifier to fine-tune the patch embedder. The refined embedder can then generate better representations in return, leading to a more accurate classifier for the next iteration. To realize more flexible and more effective embedder fine-tuning, we also introduce a teacher-student framework to efficiently distill the category knowledge in the bag classifier to help the instance-level embedder fine-tuning. Thorough experiments were conducted on four distinct datasets to validate the effectiveness of ICMIL. The experimental results consistently demonstrate that our method significantly improves the performance of existing MIL backbones, achieving state-of-the-art results. The code is available at: https://github.com/Dootmaan/ICMIL/tree/confidence_based
Iteratively Coupled Multiple Instance Learning from Instance to Bag Classifier for Whole Slide Image Classification
Mar 28, 2023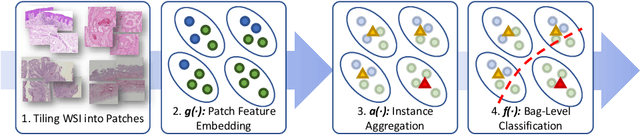
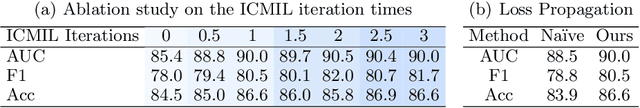
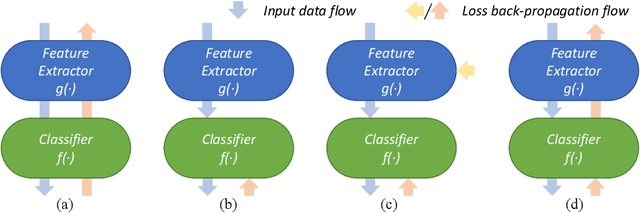
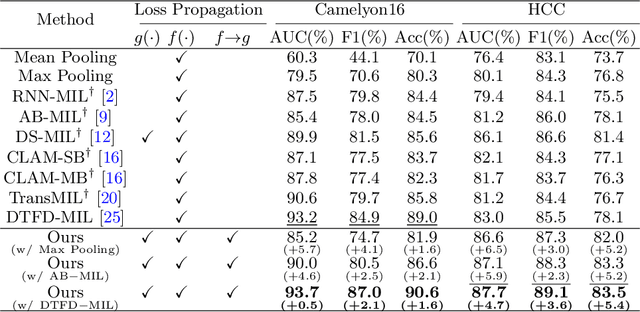
Abstract:Whole Slide Image (WSI) classification remains a challenge due to their extremely high resolution and the absence of fine-grained labels. Presently, WSIs are usually classified as a Multiple Instance Learning (MIL) problem when only slide-level labels are available. MIL methods involve a patch embedding process and a bag-level classification process, but they are prohibitively expensive to be trained end-to-end. Therefore, existing methods usually train them separately, or directly skip the training of the embedder. Such schemes hinder the patch embedder's access to slide-level labels, resulting in inconsistencies within the entire MIL pipeline. To overcome this issue, we propose a novel framework called Iteratively Coupled MIL (ICMIL), which bridges the loss back-propagation process from the bag-level classifier to the patch embedder. In ICMIL, we use category information in the bag-level classifier to guide the patch-level fine-tuning of the patch feature extractor. The refined embedder then generates better instance representations for achieving a more accurate bag-level classifier. By coupling the patch embedder and bag classifier at a low cost, our proposed framework enables information exchange between the two processes, benefiting the entire MIL classification model. We tested our framework on two datasets using three different backbones, and our experimental results demonstrate consistent performance improvements over state-of-the-art MIL methods. Code will be made available upon acceptance.
Super-Resolution Based Patch-Free 3D Medical Image Segmentation with Self-Supervised Guidance
Oct 26, 2022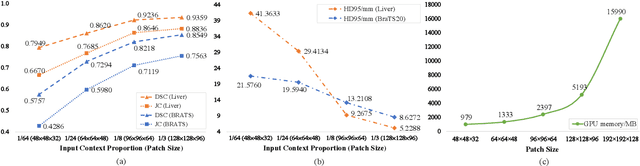
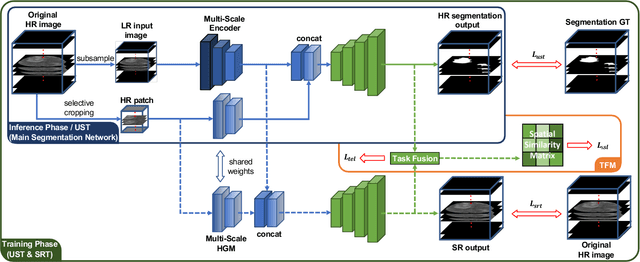
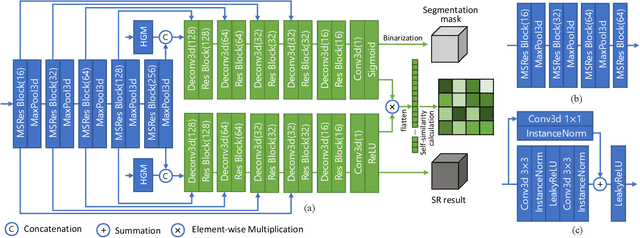
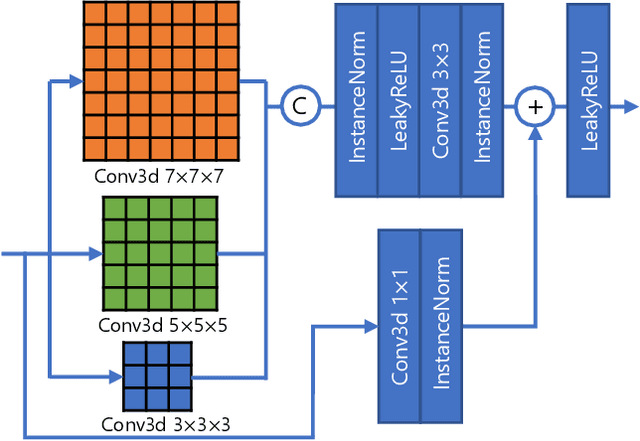
Abstract:High resolution (HR) 3D medical image segmentation plays an important role in clinical diagnoses. However, HR images are difficult to be directly processed by mainstream graphical cards due to limited video memory. Therefore, most existing 3D medical image segmentation methods use patch-based models, which ignores global context information that is useful in accurate segmentation and has low inference efficiency. To address these problems, we propose a super-resolution (SR) guided patch-free 3D medical image segmentation framework that can realize HR segmentation with global information of low-resolution (LR) input. The framework contains two tasks: semantic segmentation (main task) and super resolution (auxiliary task). To balance the information loss with the LR input, we introduce a Self-Supervised Guidance Module (SGM), which employs a selective search method to crop a HR patch from the original image as restoration guidance. Multi-scale convolutional layers are used to mitigate the scale-inconsistency between the HR guidance features and the LR features. Moreover, we propose a Task-Fusion Module (TFM) to exploit the inter connections between segmentation and SR task. This module can also be used for Test Phase Fine-tuning (TPF), leading to a better model generalization ability. When predicting, only the main segmentation task is needed, while other modules can be removed to accelerate the inference. The experiments results on two different datasets show that our framework outperforms current patch-based and patch-free models. Our model also has a four times higher inference speed compared to traditional patch-based methods. Our codes are available at: https://github.com/Dootmaan/PFSeg-Full.
Multi-phase Liver Tumor Segmentation with Spatial Aggregation and Uncertain Region Inpainting
Aug 05, 2021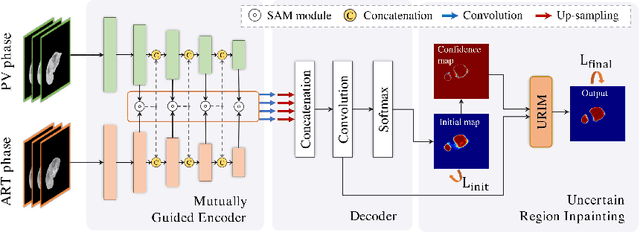
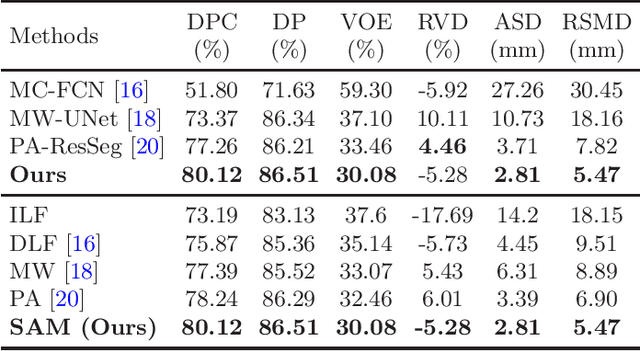
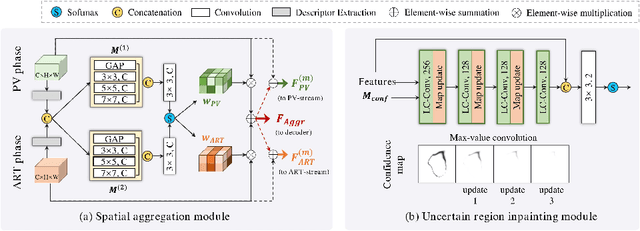
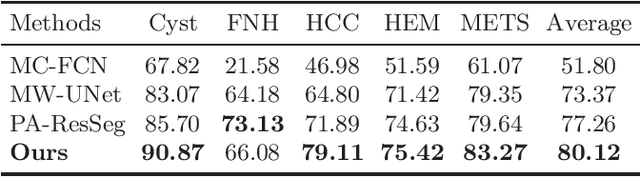
Abstract:Multi-phase computed tomography (CT) images provide crucial complementary information for accurate liver tumor segmentation (LiTS). State-of-the-art multi-phase LiTS methods usually fused cross-phase features through phase-weighted summation or channel-attention based concatenation. However, these methods ignored the spatial (pixel-wise) relationships between different phases, hence leading to insufficient feature integration. In addition, the performance of existing methods remains subject to the uncertainty in segmentation, which is particularly acute in tumor boundary regions. In this work, we propose a novel LiTS method to adequately aggregate multi-phase information and refine uncertain region segmentation. To this end, we introduce a spatial aggregation module (SAM), which encourages per-pixel interactions between different phases, to make full use of cross-phase information. Moreover, we devise an uncertain region inpainting module (URIM) to refine uncertain pixels using neighboring discriminative features. Experiments on an in-house multi-phase CT dataset of focal liver lesions (MPCT-FLLs) demonstrate that our method achieves promising liver tumor segmentation and outperforms state-of-the-arts.
PA-ResSeg: A Phase Attention Residual Network for Liver Tumor Segmentation from Multi-phase CT Images
Feb 27, 2021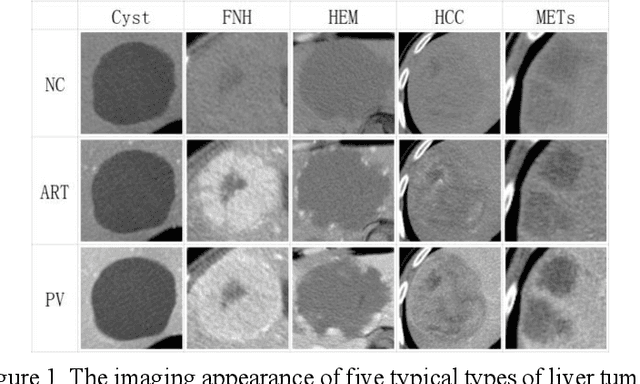
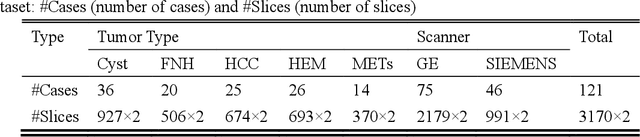
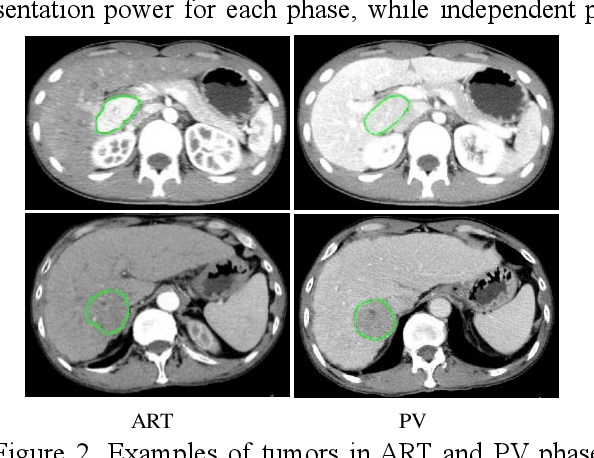
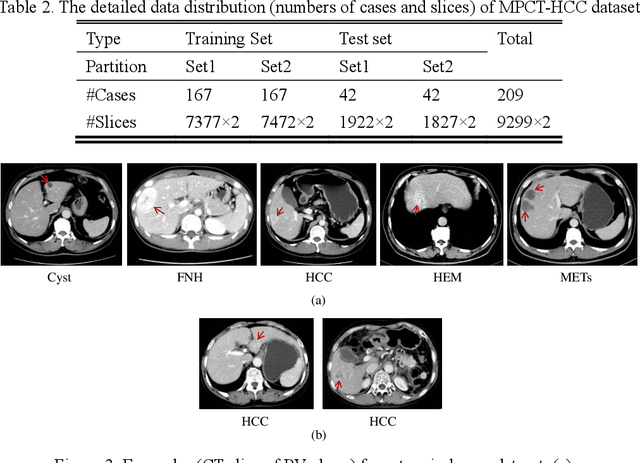
Abstract:In this paper, we propose a phase attention residual network (PA-ResSeg) to model multi-phase features for accurate liver tumor segmentation, in which a phase attention (PA) is newly proposed to additionally exploit the images of arterial (ART) phase to facilitate the segmentation of portal venous (PV) phase. The PA block consists of an intra-phase attention (Intra-PA) module and an inter-phase attention (Inter-PA) module to capture channel-wise self-dependencies and cross-phase interdependencies, respectively. Thus it enables the network to learn more representative multi-phase features by refining the PV features according to the channel dependencies and recalibrating the ART features based on the learned interdependencies between phases. We propose a PA-based multi-scale fusion (MSF) architecture to embed the PA blocks in the network at multiple levels along the encoding path to fuse multi-scale features from multi-phase images. Moreover, a 3D boundary-enhanced loss (BE-loss) is proposed for training to make the network more sensitive to boundaries. To evaluate the performance of our proposed PA-ResSeg, we conducted experiments on a multi-phase CT dataset of focal liver lesions (MPCT-FLLs). Experimental results show the effectiveness of the proposed method by achieving a dice per case (DPC) of 0.77.87, a dice global (DG) of 0.8682, a volumetric overlap error (VOE) of 0.3328 and a relative volume difference (RVD) of 0.0443 on the MPCT-FLLs. Furthermore, to validate the effectiveness and robustness of PA-ResSeg, we conducted extra experiments on another multi-phase liver tumor dataset and obtained a DPC of 0.8290, a DG of 0.9132, a VOE of 0.2637 and a RVD of 0.0163. The proposed method shows its robustness and generalization capability in different datasets and different backbones.
UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation
Apr 19, 2020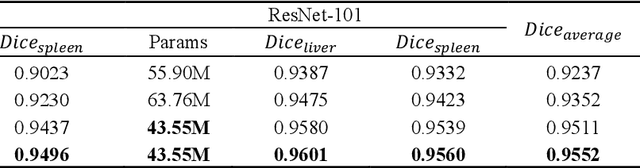
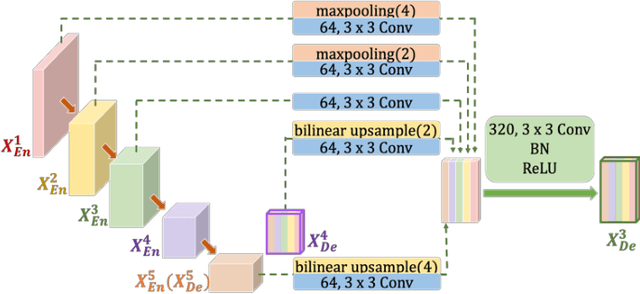
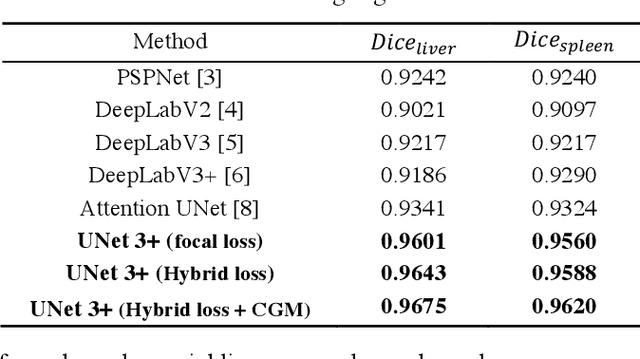
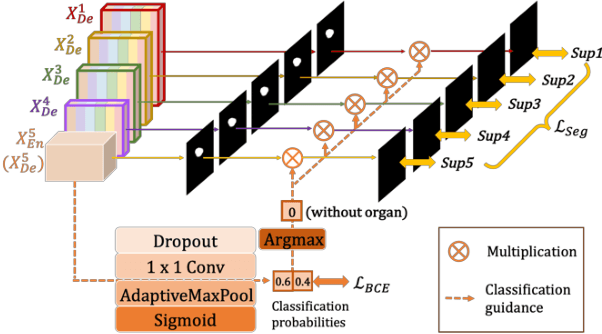
Abstract:Recently, a growing interest has been seen in deep learning-based semantic segmentation. UNet, which is one of deep learning networks with an encoder-decoder architecture, is widely used in medical image segmentation. Combining multi-scale features is one of important factors for accurate segmentation. UNet++ was developed as a modified Unet by designing an architecture with nested and dense skip connections. However, it does not explore sufficient information from full scales and there is still a large room for improvement. In this paper, we propose a novel UNet 3+, which takes advantage of full-scale skip connections and deep supervisions. The full-scale skip connections incorporate low-level details with high-level semantics from feature maps in different scales; while the deep supervision learns hierarchical representations from the full-scale aggregated feature maps. The proposed method is especially benefiting for organs that appear at varying scales. In addition to accuracy improvements, the proposed UNet 3+ can reduce the network parameters to improve the computation efficiency. We further propose a hybrid loss function and devise a classification-guided module to enhance the organ boundary and reduce the over-segmentation in a non-organ image, yielding more accurate segmentation results. The effectiveness of the proposed method is demonstrated on two datasets. The code is available at: github.com/ZJUGiveLab/UNet-Version
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge