Qingqing Chen
Exact Fit Attention in Node-Holistic Graph Convolutional Network for Improved EEG-Based Driver Fatigue Detection
Jan 25, 2025Abstract:EEG-based fatigue monitoring can effectively reduce the incidence of related traffic accidents. In the past decade, with the advancement of deep learning, convolutional neural networks (CNN) have been increasingly used for EEG signal processing. However, due to the data's non-Euclidean characteristics, existing CNNs may lose important spatial information from EEG, specifically channel correlation. Thus, we propose the node-holistic graph convolutional network (NHGNet), a model that uses graphic convolution to dynamically learn each channel's features. With exact fit attention optimization, the network captures inter-channel correlations through a trainable adjacency matrix. The interpretability is enhanced by revealing critical areas of brain activity and their interrelations in various mental states. In validations on two public datasets, NHGNet outperforms the SOTAs. Specifically, in the intra-subject, NHGNet improved detection accuracy by at least 2.34% and 3.42%, and in the inter-subjects, it improved by at least 2.09% and 15.06%. Visualization research on the model revealed that the central parietal area plays an important role in detecting fatigue levels, whereas the frontal and temporal lobes are essential for maintaining vigilance.
LSMS: Language-guided Scale-aware MedSegmentor for Medical Image Referring Segmentation
Sep 02, 2024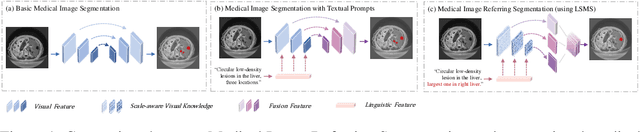
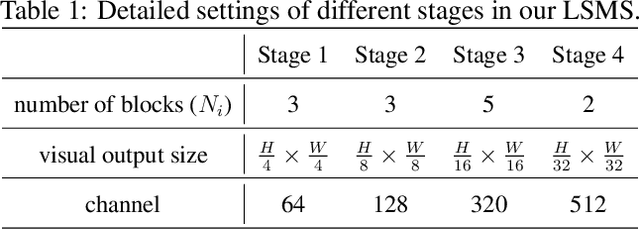
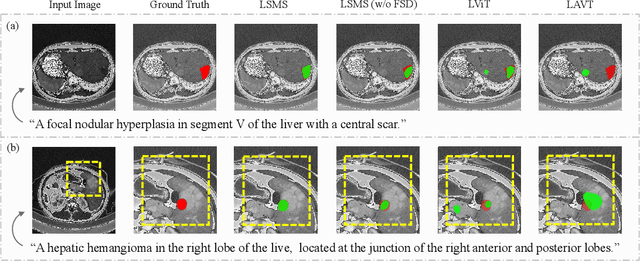
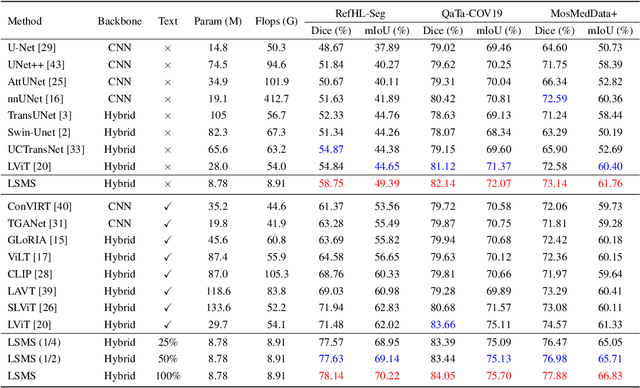
Abstract:Conventional medical image segmentation methods have been found inadequate in facilitating physicians with the identification of specific lesions for diagnosis and treatment. Given the utility of text as an instructional format, we introduce a novel task termed Medical Image Referring Segmentation (MIRS), which requires segmenting specified lesions in images based on the given language expressions. Due to the varying object scales in medical images, MIRS demands robust vision-language modeling and comprehensive multi-scale interaction for precise localization and segmentation under linguistic guidance. However, existing medical image segmentation methods fall short in meeting these demands, resulting in insufficient segmentation accuracy. In response, we propose an approach named Language-guided Scale-aware MedSegmentor (LSMS), incorporating two appealing designs: (1)~a Scale-aware Vision-Language Attention module that leverages diverse convolutional kernels to acquire rich visual knowledge and interact closely with linguistic features, thereby enhancing lesion localization capability; (2)~a Full-Scale Decoder that globally models multi-modal features across various scales, capturing complementary information between scales to accurately outline lesion boundaries. Addressing the lack of suitable datasets for MIRS, we constructed a vision-language medical dataset called Reference Hepatic Lesion Segmentation (RefHL-Seg). This dataset comprises 2,283 abdominal CT slices from 231 cases, with corresponding textual annotations and segmentation masks for various liver lesions in images. We validated the performance of LSMS for MIRS and conventional medical image segmentation tasks across various datasets. Our LSMS consistently outperforms on all datasets with lower computational costs. The code and datasets will be released.
A Dynamic Domain Adaptation Deep Learning Network for EEG-based Motor Imagery Classification
Sep 21, 2023Abstract:There is a correlation between adjacent channels of electroencephalogram (EEG), and how to represent this correlation is an issue that is currently being explored. In addition, due to inter-individual differences in EEG signals, this discrepancy results in new subjects need spend a amount of calibration time for EEG-based motor imagery brain-computer interface. In order to solve the above problems, we propose a Dynamic Domain Adaptation Based Deep Learning Network (DADL-Net). First, the EEG data is mapped to the three-dimensional geometric space and its temporal-spatial features are learned through the 3D convolution module, and then the spatial-channel attention mechanism is used to strengthen the features, and the final convolution module can further learn the spatial-temporal information of the features. Finally, to account for inter-subject and cross-sessions differences, we employ a dynamic domain-adaptive strategy, the distance between features is reduced by introducing a Maximum Mean Discrepancy loss function, and the classification layer is fine-tuned by using part of the target domain data. We verify the performance of the proposed method on BCI competition IV 2a and OpenBMI datasets. Under the intra-subject experiment, the accuracy rates of 70.42% and 73.91% were achieved on the OpenBMI and BCIC IV 2a datasets.
Super-Resolution Based Patch-Free 3D Medical Image Segmentation with Self-Supervised Guidance
Oct 26, 2022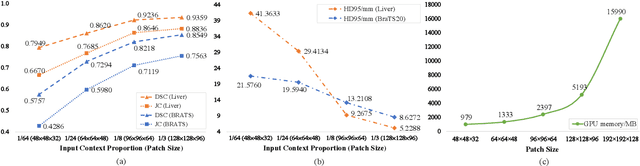
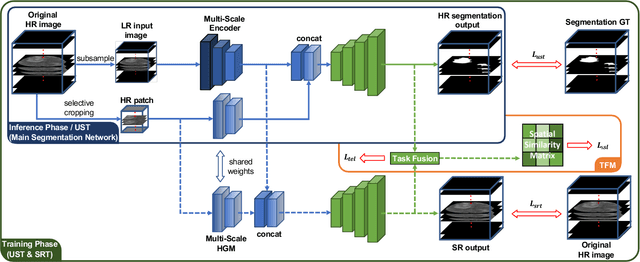
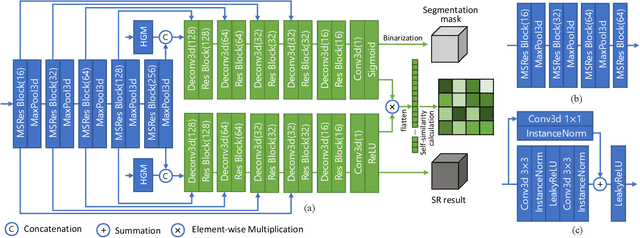
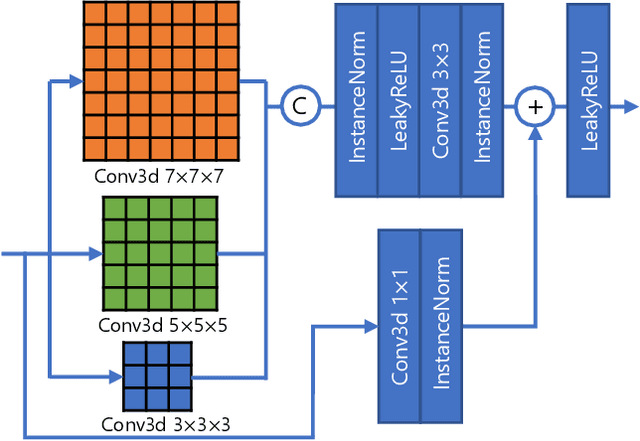
Abstract:High resolution (HR) 3D medical image segmentation plays an important role in clinical diagnoses. However, HR images are difficult to be directly processed by mainstream graphical cards due to limited video memory. Therefore, most existing 3D medical image segmentation methods use patch-based models, which ignores global context information that is useful in accurate segmentation and has low inference efficiency. To address these problems, we propose a super-resolution (SR) guided patch-free 3D medical image segmentation framework that can realize HR segmentation with global information of low-resolution (LR) input. The framework contains two tasks: semantic segmentation (main task) and super resolution (auxiliary task). To balance the information loss with the LR input, we introduce a Self-Supervised Guidance Module (SGM), which employs a selective search method to crop a HR patch from the original image as restoration guidance. Multi-scale convolutional layers are used to mitigate the scale-inconsistency between the HR guidance features and the LR features. Moreover, we propose a Task-Fusion Module (TFM) to exploit the inter connections between segmentation and SR task. This module can also be used for Test Phase Fine-tuning (TPF), leading to a better model generalization ability. When predicting, only the main segmentation task is needed, while other modules can be removed to accelerate the inference. The experiments results on two different datasets show that our framework outperforms current patch-based and patch-free models. Our model also has a four times higher inference speed compared to traditional patch-based methods. Our codes are available at: https://github.com/Dootmaan/PFSeg-Full.
Adaptively Re-weighting Multi-Loss Untrained Transformer for Sparse-View Cone-Beam CT Reconstruction
Mar 23, 2022



Abstract:Cone-Beam Computed Tomography (CBCT) has been proven useful in diagnosis, but how to shorten scanning time with lower radiation dosage and how to efficiently reconstruct 3D image remain as the main issues for clinical practice. The recent development of tomographic image reconstruction on sparse-view measurements employs deep neural networks in a supervised way to tackle such issues, whereas the success of model training requires quantity and quality of the given paired measurements/images. We propose a novel untrained Transformer to fit the CBCT inverse solver without training data. It is mainly comprised of an untrained 3D Transformer of billions of network weights and a multi-level loss function with variable weights. Unlike conventional deep neural networks (DNNs), there is no requirement of training steps in our approach. Upon observing the hardship of optimising Transformer, the variable weights within the loss function are designed to automatically update together with the iteration process, ultimately stabilising its optimisation. We evaluate the proposed approach on two publicly available datasets: SPARE and Walnut. The results show a significant performance improvement on image quality metrics with streak artefact reduction in the visualisation. We also provide a clinical report by an experienced radiologist to assess our reconstructed images in a diagnosis point of view. The source code and the optimised models are available from the corresponding author on request at the moment.
Multi-phase Liver Tumor Segmentation with Spatial Aggregation and Uncertain Region Inpainting
Aug 05, 2021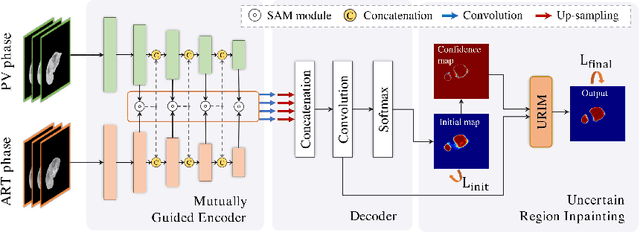
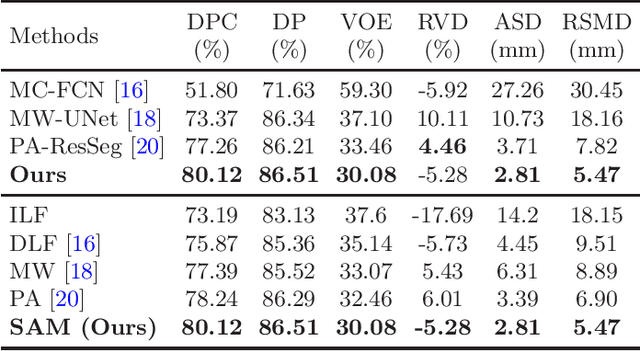
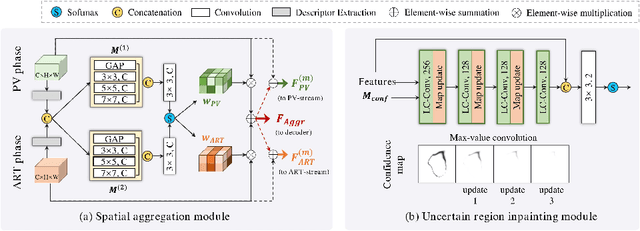
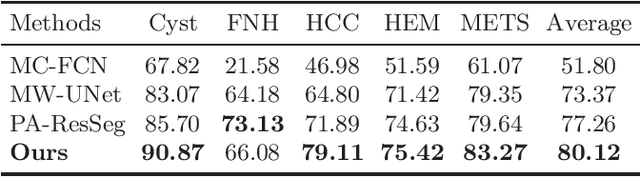
Abstract:Multi-phase computed tomography (CT) images provide crucial complementary information for accurate liver tumor segmentation (LiTS). State-of-the-art multi-phase LiTS methods usually fused cross-phase features through phase-weighted summation or channel-attention based concatenation. However, these methods ignored the spatial (pixel-wise) relationships between different phases, hence leading to insufficient feature integration. In addition, the performance of existing methods remains subject to the uncertainty in segmentation, which is particularly acute in tumor boundary regions. In this work, we propose a novel LiTS method to adequately aggregate multi-phase information and refine uncertain region segmentation. To this end, we introduce a spatial aggregation module (SAM), which encourages per-pixel interactions between different phases, to make full use of cross-phase information. Moreover, we devise an uncertain region inpainting module (URIM) to refine uncertain pixels using neighboring discriminative features. Experiments on an in-house multi-phase CT dataset of focal liver lesions (MPCT-FLLs) demonstrate that our method achieves promising liver tumor segmentation and outperforms state-of-the-arts.
PA-ResSeg: A Phase Attention Residual Network for Liver Tumor Segmentation from Multi-phase CT Images
Feb 27, 2021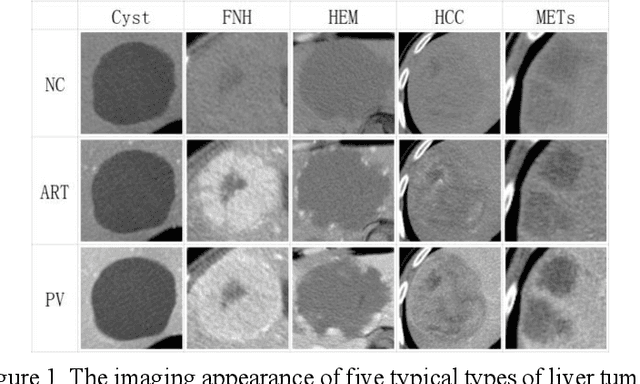
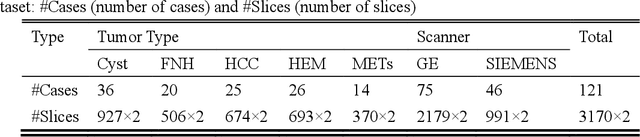
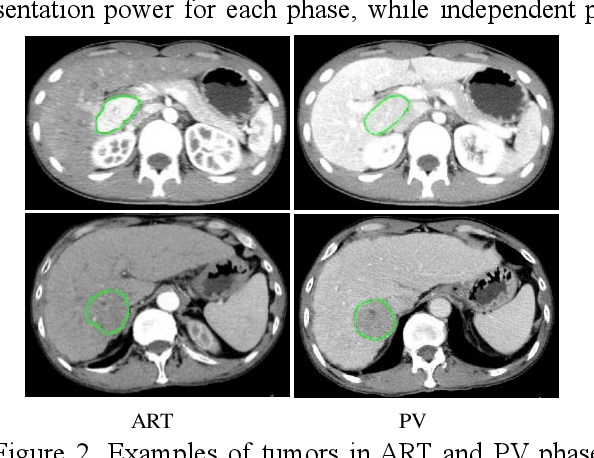
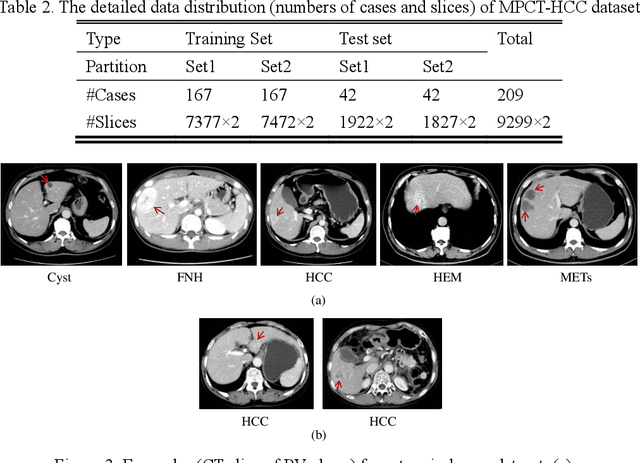
Abstract:In this paper, we propose a phase attention residual network (PA-ResSeg) to model multi-phase features for accurate liver tumor segmentation, in which a phase attention (PA) is newly proposed to additionally exploit the images of arterial (ART) phase to facilitate the segmentation of portal venous (PV) phase. The PA block consists of an intra-phase attention (Intra-PA) module and an inter-phase attention (Inter-PA) module to capture channel-wise self-dependencies and cross-phase interdependencies, respectively. Thus it enables the network to learn more representative multi-phase features by refining the PV features according to the channel dependencies and recalibrating the ART features based on the learned interdependencies between phases. We propose a PA-based multi-scale fusion (MSF) architecture to embed the PA blocks in the network at multiple levels along the encoding path to fuse multi-scale features from multi-phase images. Moreover, a 3D boundary-enhanced loss (BE-loss) is proposed for training to make the network more sensitive to boundaries. To evaluate the performance of our proposed PA-ResSeg, we conducted experiments on a multi-phase CT dataset of focal liver lesions (MPCT-FLLs). Experimental results show the effectiveness of the proposed method by achieving a dice per case (DPC) of 0.77.87, a dice global (DG) of 0.8682, a volumetric overlap error (VOE) of 0.3328 and a relative volume difference (RVD) of 0.0443 on the MPCT-FLLs. Furthermore, to validate the effectiveness and robustness of PA-ResSeg, we conducted extra experiments on another multi-phase liver tumor dataset and obtained a DPC of 0.8290, a DG of 0.9132, a VOE of 0.2637 and a RVD of 0.0163. The proposed method shows its robustness and generalization capability in different datasets and different backbones.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge