Shuyi Ouyang
Can MLLMs Guide Me Home? A Benchmark Study on Fine-Grained Visual Reasoning from Transit Maps
May 24, 2025Abstract:Multimodal large language models (MLLMs) have recently achieved significant progress in visual tasks, including semantic scene understanding and text-image alignment, with reasoning variants enhancing performance on complex tasks involving mathematics and logic. However, their capacity for reasoning tasks involving fine-grained visual understanding remains insufficiently evaluated. To address this gap, we introduce ReasonMap, a benchmark designed to assess the fine-grained visual understanding and spatial reasoning abilities of MLLMs. ReasonMap encompasses high-resolution transit maps from 30 cities across 13 countries and includes 1,008 question-answer pairs spanning two question types and three templates. Furthermore, we design a two-level evaluation pipeline that properly assesses answer correctness and quality. Comprehensive evaluations of 15 popular MLLMs, including both base and reasoning variants, reveal a counterintuitive pattern: among open-source models, base models outperform reasoning ones, while the opposite trend is observed in closed-source models. Additionally, performance generally degrades when visual inputs are masked, indicating that while MLLMs can leverage prior knowledge to answer some questions, fine-grained visual reasoning tasks still require genuine visual perception for strong performance. Our benchmark study offers new insights into visual reasoning and contributes to investigating the gap between open-source and closed-source models.
SemSim: Revisiting Weak-to-Strong Consistency from a Semantic Similarity Perspective for Semi-supervised Medical Image Segmentation
Oct 17, 2024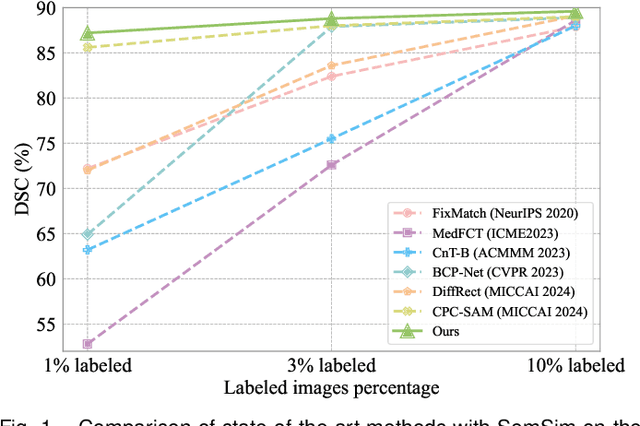
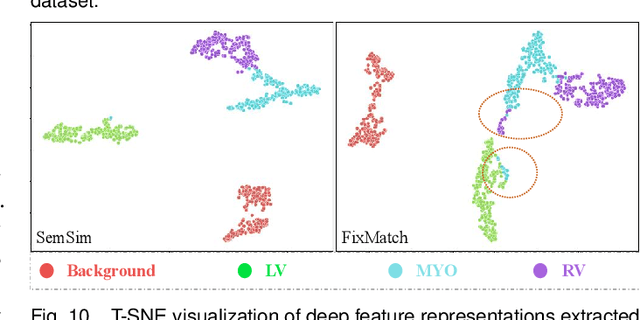
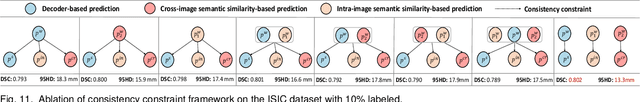
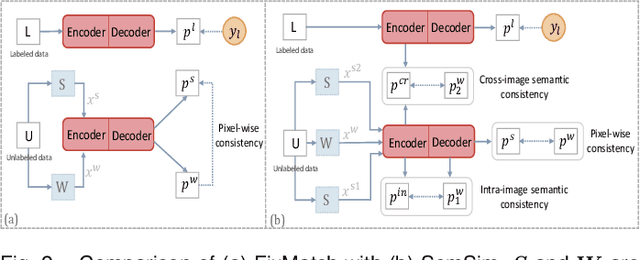
Abstract:Semi-supervised learning (SSL) for medical image segmentation is a challenging yet highly practical task, which reduces reliance on large-scale labeled dataset by leveraging unlabeled samples. Among SSL techniques, the weak-to-strong consistency framework, popularized by FixMatch, has emerged as a state-of-the-art method in classification tasks. Notably, such a simple pipeline has also shown competitive performance in medical image segmentation. However, two key limitations still persist, impeding its efficient adaptation: (1) the neglect of contextual dependencies results in inconsistent predictions for similar semantic features, leading to incomplete object segmentation; (2) the lack of exploitation of semantic similarity between labeled and unlabeled data induces considerable class-distribution discrepancy. To address these limitations, we propose a novel semi-supervised framework based on FixMatch, named SemSim, powered by two appealing designs from semantic similarity perspective: (1) rectifying pixel-wise prediction by reasoning about the intra-image pair-wise affinity map, thus integrating contextual dependencies explicitly into the final prediction; (2) bridging labeled and unlabeled data via a feature querying mechanism for compact class representation learning, which fully considers cross-image anatomical similarities. As the reliable semantic similarity extraction depends on robust features, we further introduce an effective spatial-aware fusion module (SFM) to explore distinctive information from multiple scales. Extensive experiments show that SemSim yields consistent improvements over the state-of-the-art methods across three public segmentation benchmarks.
LSMS: Language-guided Scale-aware MedSegmentor for Medical Image Referring Segmentation
Sep 02, 2024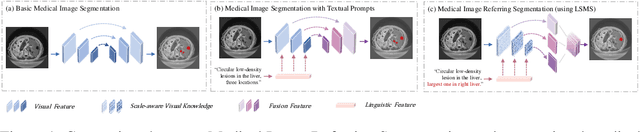
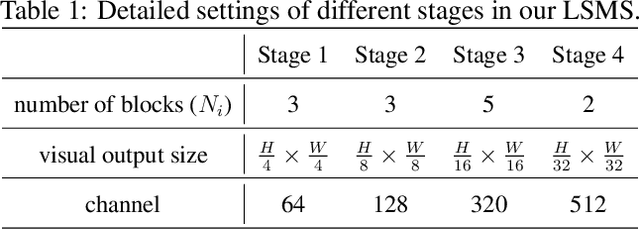
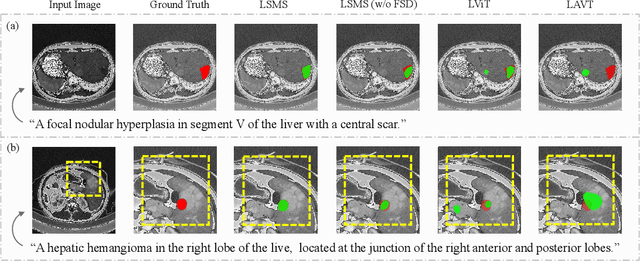
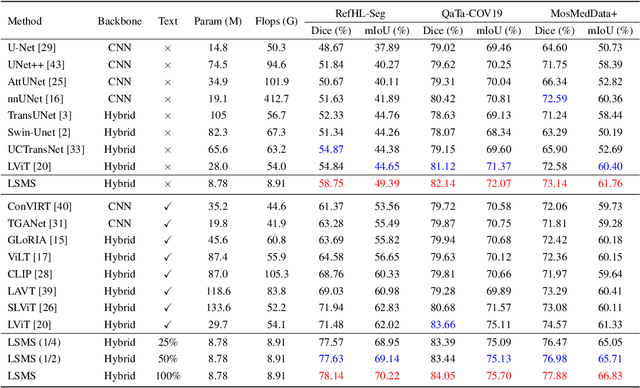
Abstract:Conventional medical image segmentation methods have been found inadequate in facilitating physicians with the identification of specific lesions for diagnosis and treatment. Given the utility of text as an instructional format, we introduce a novel task termed Medical Image Referring Segmentation (MIRS), which requires segmenting specified lesions in images based on the given language expressions. Due to the varying object scales in medical images, MIRS demands robust vision-language modeling and comprehensive multi-scale interaction for precise localization and segmentation under linguistic guidance. However, existing medical image segmentation methods fall short in meeting these demands, resulting in insufficient segmentation accuracy. In response, we propose an approach named Language-guided Scale-aware MedSegmentor (LSMS), incorporating two appealing designs: (1)~a Scale-aware Vision-Language Attention module that leverages diverse convolutional kernels to acquire rich visual knowledge and interact closely with linguistic features, thereby enhancing lesion localization capability; (2)~a Full-Scale Decoder that globally models multi-modal features across various scales, capturing complementary information between scales to accurately outline lesion boundaries. Addressing the lack of suitable datasets for MIRS, we constructed a vision-language medical dataset called Reference Hepatic Lesion Segmentation (RefHL-Seg). This dataset comprises 2,283 abdominal CT slices from 231 cases, with corresponding textual annotations and segmentation masks for various liver lesions in images. We validated the performance of LSMS for MIRS and conventional medical image segmentation tasks across various datasets. Our LSMS consistently outperforms on all datasets with lower computational costs. The code and datasets will be released.
HSVLT: Hierarchical Scale-Aware Vision-Language Transformer for Multi-Label Image Classification
Jul 23, 2024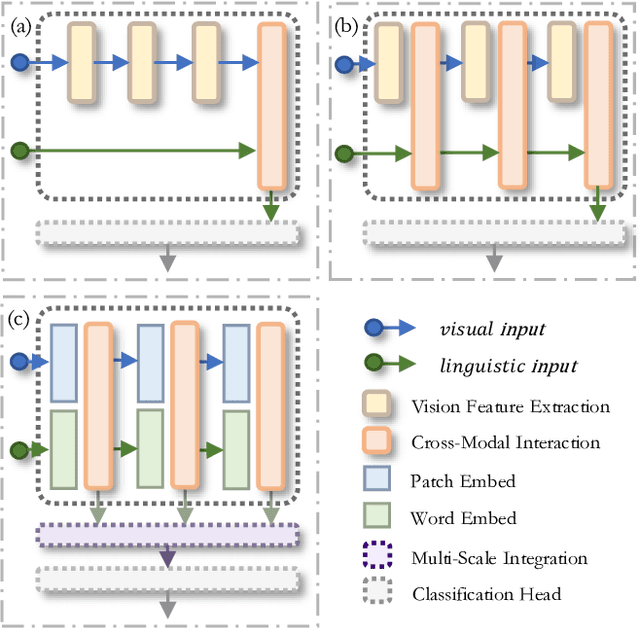
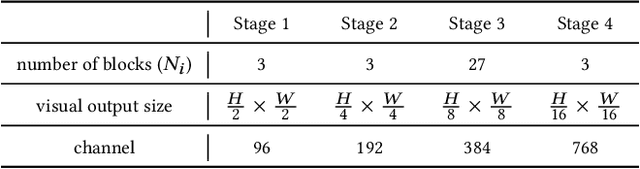
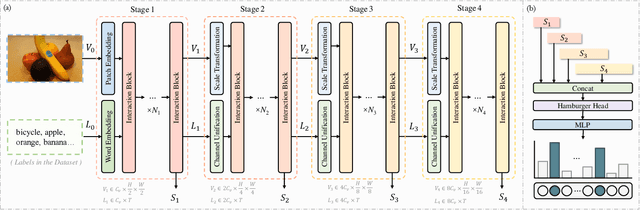
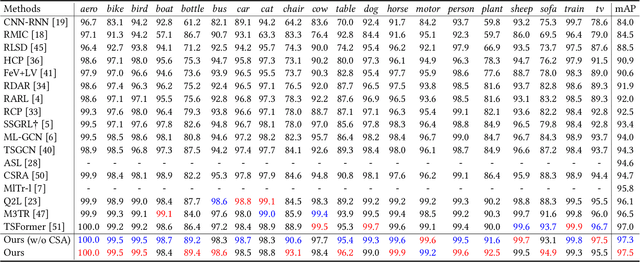
Abstract:The task of multi-label image classification involves recognizing multiple objects within a single image. Considering both valuable semantic information contained in the labels and essential visual features presented in the image, tight visual-linguistic interactions play a vital role in improving classification performance. Moreover, given the potential variance in object size and appearance within a single image, attention to features of different scales can help to discover possible objects in the image. Recently, Transformer-based methods have achieved great success in multi-label image classification by leveraging the advantage of modeling long-range dependencies, but they have several limitations. Firstly, existing methods treat visual feature extraction and cross-modal fusion as separate steps, resulting in insufficient visual-linguistic alignment in the joint semantic space. Additionally, they only extract visual features and perform cross-modal fusion at a single scale, neglecting objects with different characteristics. To address these issues, we propose a Hierarchical Scale-Aware Vision-Language Transformer (HSVLT) with two appealing designs: (1)~A hierarchical multi-scale architecture that involves a Cross-Scale Aggregation module, which leverages joint multi-modal features extracted from multiple scales to recognize objects of varying sizes and appearances in images. (2)~Interactive Visual-Linguistic Attention, a novel attention mechanism module that tightly integrates cross-modal interaction, enabling the joint updating of visual, linguistic and multi-modal features. We have evaluated our method on three benchmark datasets. The experimental results demonstrate that HSVLT surpasses state-of-the-art methods with lower computational cost.
* 10 pages, 6 figures
A Survey on Domain Generalization for Medical Image Analysis
Feb 13, 2024Abstract:Medical Image Analysis (MedIA) has emerged as a crucial tool in computer-aided diagnosis systems, particularly with the advancement of deep learning (DL) in recent years. However, well-trained deep models often experience significant performance degradation when deployed in different medical sites, modalities, and sequences, known as a domain shift issue. In light of this, Domain Generalization (DG) for MedIA aims to address the domain shift challenge by generalizing effectively and performing robustly across unknown data distributions. This paper presents the a comprehensive review of substantial developments in this area. First, we provide a formal definition of domain shift and domain generalization in medical field, and discuss several related settings. Subsequently, we summarize the recent methods from three viewpoints: data manipulation level, feature representation level, and model training level, and present some algorithms in detail for each viewpoints. Furthermore, we introduce the commonly used datasets. Finally, we summarize existing literature and present some potential research topics for the future. For this survey, we also created a GitHub project by collecting the supporting resources, at the link: https://github.com/Ziwei-Niu/DG_for_MedIA
M2ORT: Many-To-One Regression Transformer for Spatial Transcriptomics Prediction from Histopathology Images
Jan 24, 2024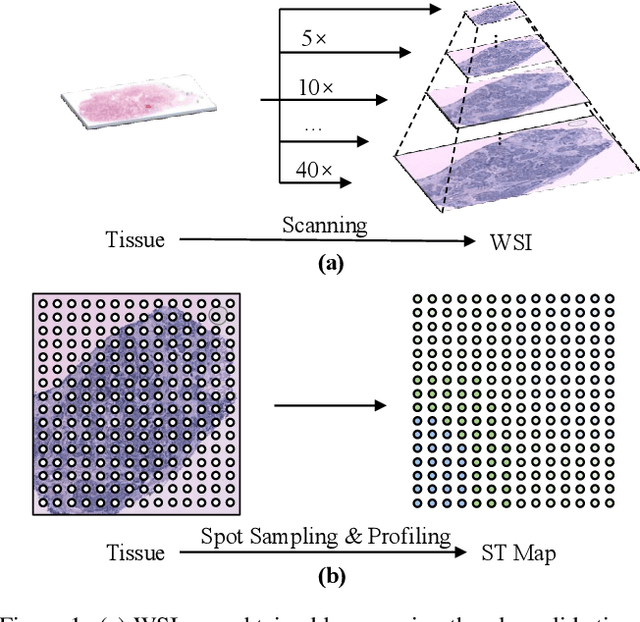
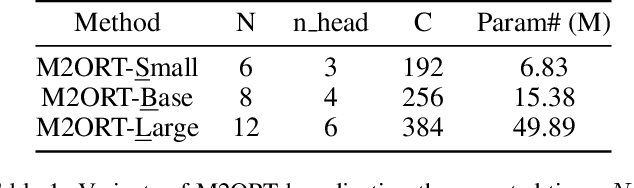
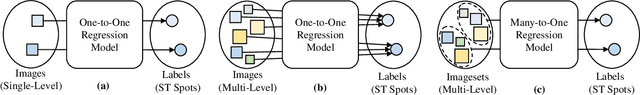
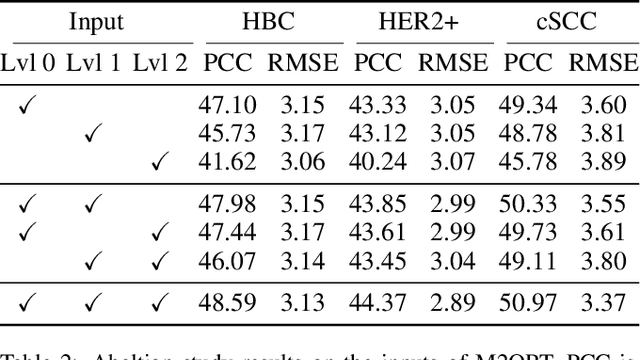
Abstract:The advancement of Spatial Transcriptomics (ST) has facilitated the spatially-aware profiling of gene expressions based on histopathology images. Although ST data offers valuable insights into the micro-environment of tumors, its acquisition cost remains expensive. Therefore, directly predicting the ST expressions from digital pathology images is desired. Current methods usually adopt existing regression backbones for this task, which ignore the inherent multi-scale hierarchical data structure of digital pathology images. To address this limit, we propose M2ORT, a many-to-one regression Transformer that can accommodate the hierarchical structure of the pathology images through a decoupled multi-scale feature extractor. Different from traditional models that are trained with one-to-one image-label pairs, M2ORT accepts multiple pathology images of different magnifications at a time to jointly predict the gene expressions at their corresponding common ST spot, aiming at learning a many-to-one relationship through training. We have tested M2ORT on three public ST datasets and the experimental results show that M2ORT can achieve state-of-the-art performance with fewer parameters and floating-point operations (FLOPs). The code is available at: https://github.com/Dootmaan/M2ORT/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge