Yutaro Iwamoto
Accurate Tracking of Arabidopsis Root Cortex Cell Nuclei in 3D Time-Lapse Microscopy Images Based on Genetic Algorithm
Apr 17, 2025Abstract:Arabidopsis is a widely used model plant to gain basic knowledge on plant physiology and development. Live imaging is an important technique to visualize and quantify elemental processes in plant development. To uncover novel theories underlying plant growth and cell division, accurate cell tracking on live imaging is of utmost importance. The commonly used cell tracking software, TrackMate, adopts tracking-by-detection fashion, which applies Laplacian of Gaussian (LoG) for blob detection, and Linear Assignment Problem (LAP) tracker for tracking. However, they do not perform sufficiently when cells are densely arranged. To alleviate the problems mentioned above, we propose an accurate tracking method based on Genetic algorithm (GA) using knowledge of Arabidopsis root cellular patterns and spatial relationship among volumes. Our method can be described as a coarse-to-fine method, in which we first conducted relatively easy line-level tracking of cell nuclei, then performed complicated nuclear tracking based on known linear arrangement of cell files and their spatial relationship between nuclei. Our method has been evaluated on a long-time live imaging dataset of Arabidopsis root tips, and with minor manual rectification, it accurately tracks nuclei. To the best of our knowledge, this research represents the first successful attempt to address a long-standing problem in the field of time-lapse microscopy in the root meristem by proposing an accurate tracking method for Arabidopsis root nuclei.
Super-Resolution Based Patch-Free 3D Medical Image Segmentation with Self-Supervised Guidance
Oct 26, 2022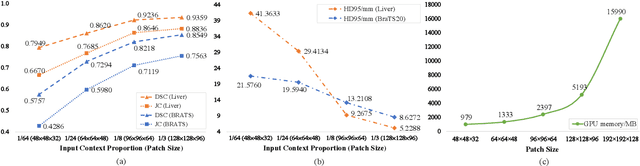
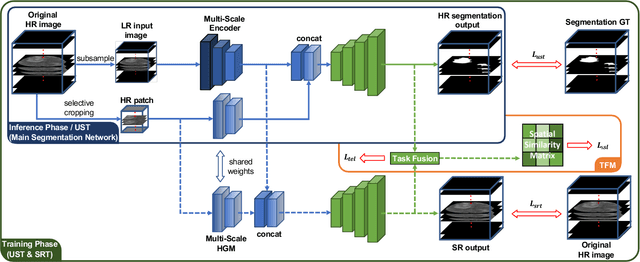
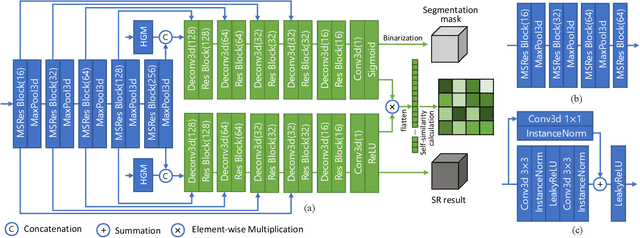
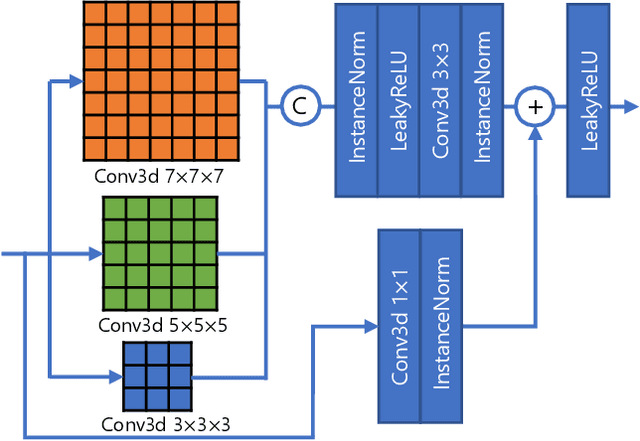
Abstract:High resolution (HR) 3D medical image segmentation plays an important role in clinical diagnoses. However, HR images are difficult to be directly processed by mainstream graphical cards due to limited video memory. Therefore, most existing 3D medical image segmentation methods use patch-based models, which ignores global context information that is useful in accurate segmentation and has low inference efficiency. To address these problems, we propose a super-resolution (SR) guided patch-free 3D medical image segmentation framework that can realize HR segmentation with global information of low-resolution (LR) input. The framework contains two tasks: semantic segmentation (main task) and super resolution (auxiliary task). To balance the information loss with the LR input, we introduce a Self-Supervised Guidance Module (SGM), which employs a selective search method to crop a HR patch from the original image as restoration guidance. Multi-scale convolutional layers are used to mitigate the scale-inconsistency between the HR guidance features and the LR features. Moreover, we propose a Task-Fusion Module (TFM) to exploit the inter connections between segmentation and SR task. This module can also be used for Test Phase Fine-tuning (TPF), leading to a better model generalization ability. When predicting, only the main segmentation task is needed, while other modules can be removed to accelerate the inference. The experiments results on two different datasets show that our framework outperforms current patch-based and patch-free models. Our model also has a four times higher inference speed compared to traditional patch-based methods. Our codes are available at: https://github.com/Dootmaan/PFSeg-Full.
ScaleFormer: Revisiting the Transformer-based Backbones from a Scale-wise Perspective for Medical Image Segmentation
Jul 29, 2022



Abstract:Recently, a variety of vision transformers have been developed as their capability of modeling long-range dependency. In current transformer-based backbones for medical image segmentation, convolutional layers were replaced with pure transformers, or transformers were added to the deepest encoder to learn global context. However, there are mainly two challenges in a scale-wise perspective: (1) intra-scale problem: the existing methods lacked in extracting local-global cues in each scale, which may impact the signal propagation of small objects; (2) inter-scale problem: the existing methods failed to explore distinctive information from multiple scales, which may hinder the representation learning from objects with widely variable size, shape and location. To address these limitations, we propose a novel backbone, namely ScaleFormer, with two appealing designs: (1) A scale-wise intra-scale transformer is designed to couple the CNN-based local features with the transformer-based global cues in each scale, where the row-wise and column-wise global dependencies can be extracted by a lightweight Dual-Axis MSA. (2) A simple and effective spatial-aware inter-scale transformer is designed to interact among consensual regions in multiple scales, which can highlight the cross-scale dependency and resolve the complex scale variations. Experimental results on different benchmarks demonstrate that our Scale-Former outperforms the current state-of-the-art methods. The code is publicly available at: https://github.com/ZJUGiveLab/ScaleFormer.
Efficient and Accurate Hyperspectral Pansharpening Using 3D VolumeNet and 2.5D Texture Transfer
Mar 08, 2022


Abstract:Recently, convolutional neural networks (CNN) have obtained promising results in single-image SR for hyperspectral pansharpening. However, enhancing CNNs' representation ability with fewer parameters and a shorter prediction time is a challenging and critical task. In this paper, we propose a novel multi-spectral image fusion method using a combination of the previously proposed 3D CNN model VolumeNet and 2.5D texture transfer method using other modality high resolution (HR) images. Since a multi-spectral (MS) image consists of several bands and each band is a 2D image slice, MS images can be seen as 3D data. Thus, we use the previously proposed VolumeNet to fuse HR panchromatic (PAN) images and bicubic interpolated MS images. Because the proposed 3D VolumeNet can effectively improve the accuracy by expanding the receptive field of the model, and due to its lightweight structure, we can achieve better performance against the existing method without purchasing a large number of remote sensing images for training. In addition, VolumeNet can restore the high-frequency information lost in the HR MR image as much as possible, reducing the difficulty of feature extraction in the following step: 2.5D texture transfer. As one of the latest technologies, deep learning-based texture transfer has been demonstrated to effectively and efficiently improve the visual performance and quality evaluation indicators of image reconstruction. Different from the texture transfer processing of RGB image, we use HR PAN images as the reference images and perform texture transfer for each frequency band of MS images, which is named 2.5D texture transfer. The experimental results show that the proposed method outperforms the existing methods in terms of objective accuracy assessment, method efficiency, and visual subjective evaluation.
Mixed Transformer U-Net For Medical Image Segmentation
Nov 11, 2021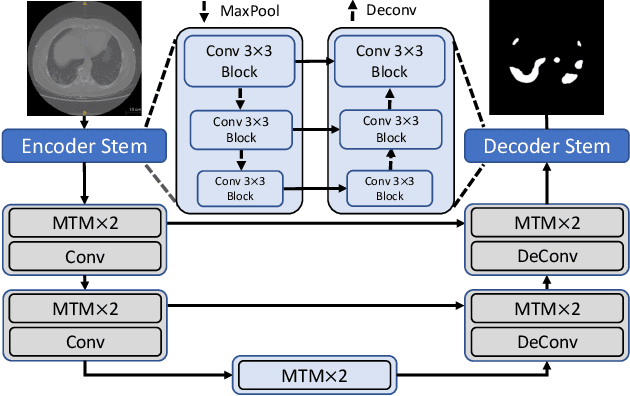
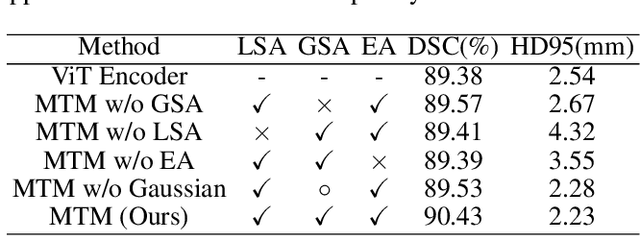
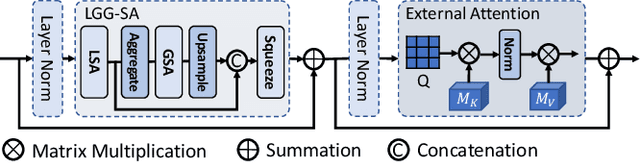
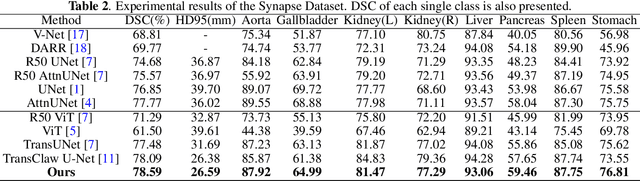
Abstract:Though U-Net has achieved tremendous success in medical image segmentation tasks, it lacks the ability to explicitly model long-range dependencies. Therefore, Vision Transformers have emerged as alternative segmentation structures recently, for their innate ability of capturing long-range correlations through Self-Attention (SA). However, Transformers usually rely on large-scale pre-training and have high computational complexity. Furthermore, SA can only model self-affinities within a single sample, ignoring the potential correlations of the overall dataset. To address these problems, we propose a novel Transformer module named Mixed Transformer Module (MTM) for simultaneous inter- and intra- affinities learning. MTM first calculates self-affinities efficiently through our well-designed Local-Global Gaussian-Weighted Self-Attention (LGG-SA). Then, it mines inter-connections between data samples through External Attention (EA). By using MTM, we construct a U-shaped model named Mixed Transformer U-Net (MT-UNet) for accurate medical image segmentation. We test our method on two different public datasets, and the experimental results show that the proposed method achieves better performance over other state-of-the-art methods. The code is available at: https://github.com/Dootmaan/MT-UNet.
Genotype-Guided Radiomics Signatures for Recurrence Prediction of Non-Small-Cell Lung Cancer
Apr 29, 2021



Abstract:Non-small cell lung cancer (NSCLC) is a serious disease and has a high recurrence rate after the surgery. Recently, many machine learning methods have been proposed for recurrence prediction. The methods using gene data have high prediction accuracy but require high cost. Although the radiomics signatures using only CT image are not expensive, its accuracy is relatively low. In this paper, we propose a genotype-guided radiomics method (GGR) for obtaining high prediction accuracy with low cost. We used a public radiogenomics dataset of NSCLC, which includes CT images and gene data. The proposed method is a two-step method, which consists of two models. The first model is a gene estimation model, which is used to estimate the gene expression from radiomics features and deep features extracted from computer tomography (CT) image. The second model is used to predict the recurrence using the estimated gene expression data. The proposed GGR method designed based on hybrid features which is combination of handcrafted-based and deep learning-based. The experiments demonstrated that the prediction accuracy can be improved significantly from 78.61% (existing radiomics method) and 79.14% (deep learning method) to 83.28% by the proposed GGR.
Graph-based Pyramid Global Context Reasoning with a Saliency-aware Projection for COVID-19 Lung Infections Segmentation
Mar 07, 2021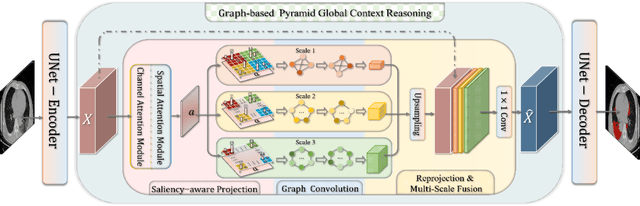
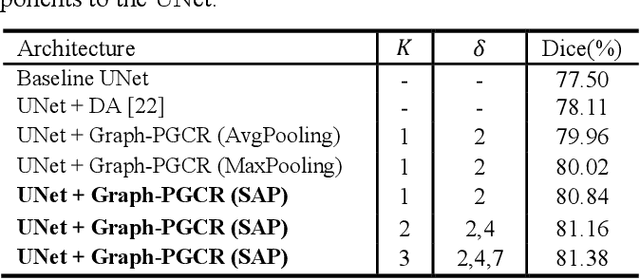
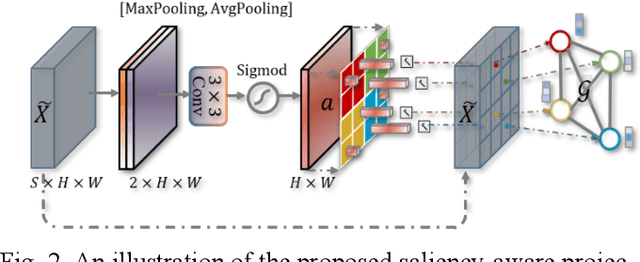
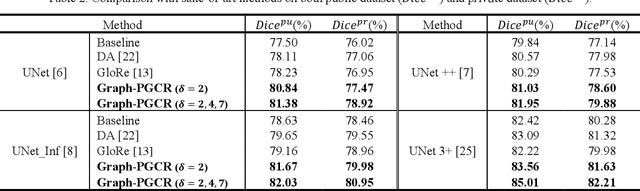
Abstract:Coronavirus Disease 2019 (COVID-19) has rapidly spread in 2020, emerging a mass of studies for lung infection segmentation from CT images. Though many methods have been proposed for this issue, it is a challenging task because of infections of various size appearing in different lobe zones. To tackle these issues, we propose a Graph-based Pyramid Global Context Reasoning (Graph-PGCR) module, which is capable of modeling long-range dependencies among disjoint infections as well as adapt size variation. We first incorporate graph convolution to exploit long-term contextual information from multiple lobe zones. Different from previous average pooling or maximum object probability, we propose a saliency-aware projection mechanism to pick up infection-related pixels as a set of graph nodes. After graph reasoning, the relation-aware features are reversed back to the original coordinate space for the down-stream tasks. We further construct multiple graphs with different sampling rates to handle the size variation problem. To this end, distinct multi-scale long-range contextual patterns can be captured. Our Graph-PGCR module is plug-and-play, which can be integrated into any architecture to improve its performance. Experiments demonstrated that the proposed method consistently boost the performance of state-of-the-art backbone architectures on both of public and our private COVID-19 datasets.
PA-ResSeg: A Phase Attention Residual Network for Liver Tumor Segmentation from Multi-phase CT Images
Feb 27, 2021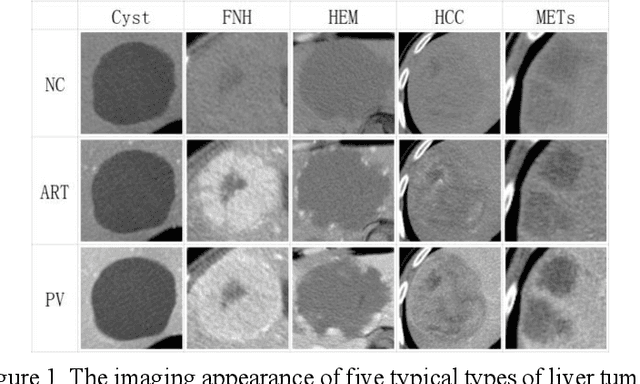
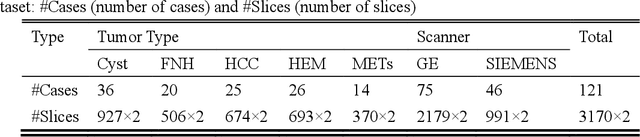
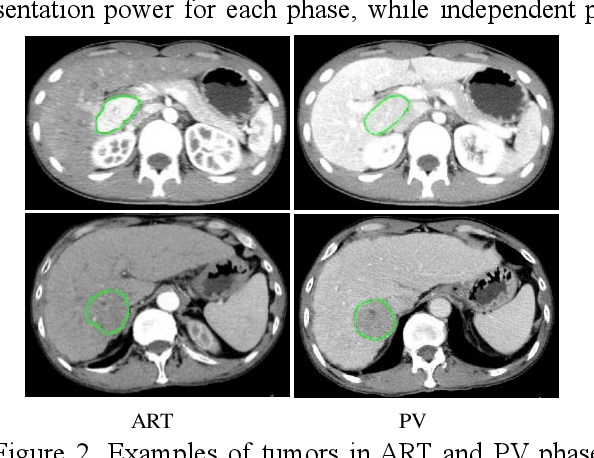
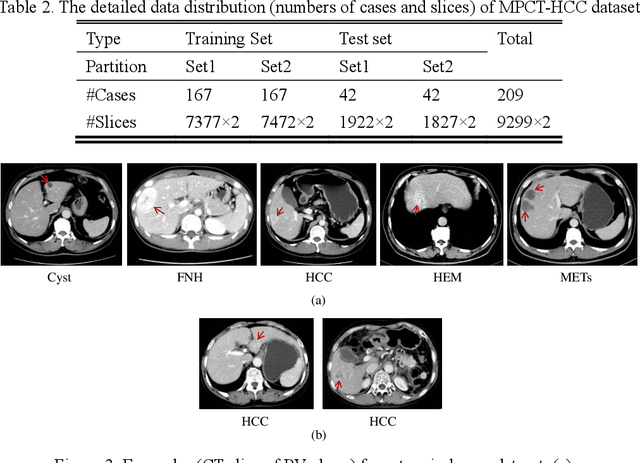
Abstract:In this paper, we propose a phase attention residual network (PA-ResSeg) to model multi-phase features for accurate liver tumor segmentation, in which a phase attention (PA) is newly proposed to additionally exploit the images of arterial (ART) phase to facilitate the segmentation of portal venous (PV) phase. The PA block consists of an intra-phase attention (Intra-PA) module and an inter-phase attention (Inter-PA) module to capture channel-wise self-dependencies and cross-phase interdependencies, respectively. Thus it enables the network to learn more representative multi-phase features by refining the PV features according to the channel dependencies and recalibrating the ART features based on the learned interdependencies between phases. We propose a PA-based multi-scale fusion (MSF) architecture to embed the PA blocks in the network at multiple levels along the encoding path to fuse multi-scale features from multi-phase images. Moreover, a 3D boundary-enhanced loss (BE-loss) is proposed for training to make the network more sensitive to boundaries. To evaluate the performance of our proposed PA-ResSeg, we conducted experiments on a multi-phase CT dataset of focal liver lesions (MPCT-FLLs). Experimental results show the effectiveness of the proposed method by achieving a dice per case (DPC) of 0.77.87, a dice global (DG) of 0.8682, a volumetric overlap error (VOE) of 0.3328 and a relative volume difference (RVD) of 0.0443 on the MPCT-FLLs. Furthermore, to validate the effectiveness and robustness of PA-ResSeg, we conducted extra experiments on another multi-phase liver tumor dataset and obtained a DPC of 0.8290, a DG of 0.9132, a VOE of 0.2637 and a RVD of 0.0163. The proposed method shows its robustness and generalization capability in different datasets and different backbones.
VolumeNet: A Lightweight Parallel Network for Super-Resolution of Medical Volumetric Data
Oct 24, 2020



Abstract:Deep learning-based super-resolution (SR) techniques have generally achieved excellent performance in the computer vision field. Recently, it has been proven that three-dimensional (3D) SR for medical volumetric data delivers better visual results than conventional two-dimensional (2D) processing. However, deepening and widening 3D networks increases training difficulty significantly due to the large number of parameters and small number of training samples. Thus, we propose a 3D convolutional neural network (CNN) for SR of medical volumetric data called ParallelNet using parallel connections. We construct a parallel connection structure based on the group convolution and feature aggregation to build a 3D CNN that is as wide as possible with few parameters. As a result, the model thoroughly learns more feature maps with larger receptive fields. In addition, to further improve accuracy, we present an efficient version of ParallelNet (called VolumeNet), which reduces the number of parameters and deepens ParallelNet using a proposed lightweight building block module called the Queue module. Unlike most lightweight CNNs based on depthwise convolutions, the Queue module is primarily constructed using separable 2D cross-channel convolutions. As a result, the number of network parameters and computational complexity can be reduced significantly while maintaining accuracy due to full channel fusion. Experimental results demonstrate that the proposed VolumeNet significantly reduces the number of model parameters and achieves high precision results compared to state-of-the-art methods.
Unsupervised MRI Super-Resolution Using Deep External Learning and Guided Residual Dense Network with Multimodal Image Priors
Oct 02, 2020



Abstract:Deep learning techniques have led to state-of-the-art single image super-resolution (SISR) with natural images. Pairs of high-resolution (HR) and low-resolution (LR) images are used to train the deep learning model (mapping function). These techniques have also been applied to medical image super-resolution (SR). Compared with natural images, medical images have several unique characteristics. First, there are no HR images for training in real clinical applications because of the limitations of imaging systems and clinical requirements. Second, other modal HR images are available (e.g., HR T1-weighted images are available for enhancing LR T2-weighted images). In this paper, we propose an unsupervised SISR technique based on simple prior knowledge of the human anatomy; this technique does not require HR images for training. Furthermore, we present a guided residual dense network, which incorporates a residual dense network with a guided deep convolutional neural network for enhancing the resolution of LR images by referring to different HR images of the same subject. Experiments on a publicly available brain MRI database showed that our proposed method achieves better performance than the state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge