Xianhua Han
ScaleFormer: Revisiting the Transformer-based Backbones from a Scale-wise Perspective for Medical Image Segmentation
Jul 29, 2022



Abstract:Recently, a variety of vision transformers have been developed as their capability of modeling long-range dependency. In current transformer-based backbones for medical image segmentation, convolutional layers were replaced with pure transformers, or transformers were added to the deepest encoder to learn global context. However, there are mainly two challenges in a scale-wise perspective: (1) intra-scale problem: the existing methods lacked in extracting local-global cues in each scale, which may impact the signal propagation of small objects; (2) inter-scale problem: the existing methods failed to explore distinctive information from multiple scales, which may hinder the representation learning from objects with widely variable size, shape and location. To address these limitations, we propose a novel backbone, namely ScaleFormer, with two appealing designs: (1) A scale-wise intra-scale transformer is designed to couple the CNN-based local features with the transformer-based global cues in each scale, where the row-wise and column-wise global dependencies can be extracted by a lightweight Dual-Axis MSA. (2) A simple and effective spatial-aware inter-scale transformer is designed to interact among consensual regions in multiple scales, which can highlight the cross-scale dependency and resolve the complex scale variations. Experimental results on different benchmarks demonstrate that our Scale-Former outperforms the current state-of-the-art methods. The code is publicly available at: https://github.com/ZJUGiveLab/ScaleFormer.
UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation
Apr 19, 2020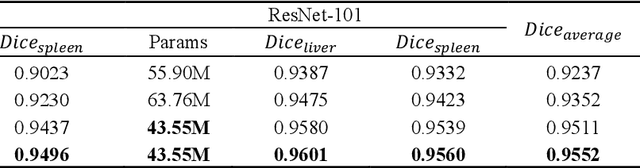
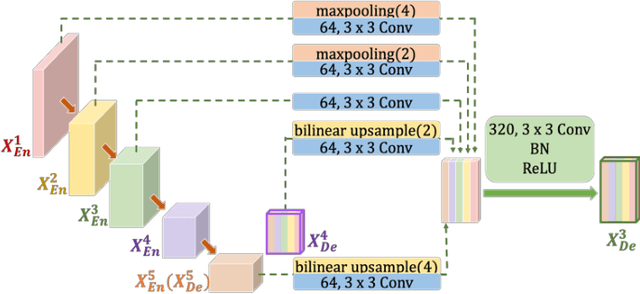
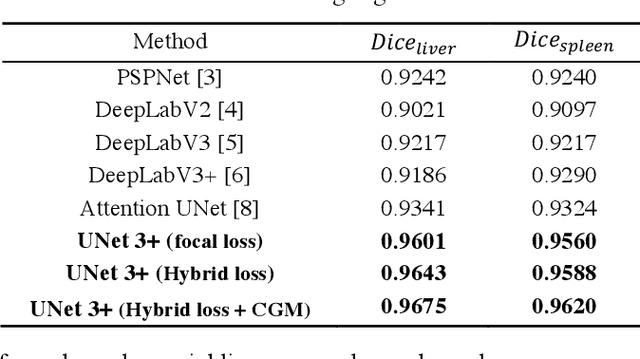
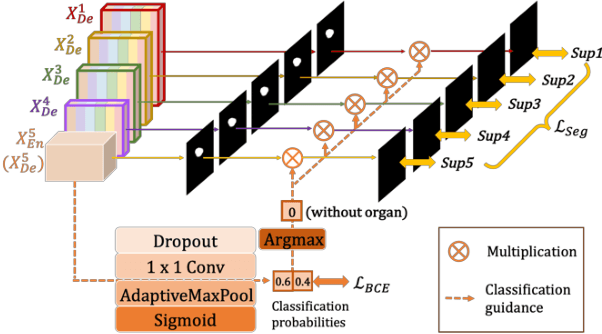
Abstract:Recently, a growing interest has been seen in deep learning-based semantic segmentation. UNet, which is one of deep learning networks with an encoder-decoder architecture, is widely used in medical image segmentation. Combining multi-scale features is one of important factors for accurate segmentation. UNet++ was developed as a modified Unet by designing an architecture with nested and dense skip connections. However, it does not explore sufficient information from full scales and there is still a large room for improvement. In this paper, we propose a novel UNet 3+, which takes advantage of full-scale skip connections and deep supervisions. The full-scale skip connections incorporate low-level details with high-level semantics from feature maps in different scales; while the deep supervision learns hierarchical representations from the full-scale aggregated feature maps. The proposed method is especially benefiting for organs that appear at varying scales. In addition to accuracy improvements, the proposed UNet 3+ can reduce the network parameters to improve the computation efficiency. We further propose a hybrid loss function and devise a classification-guided module to enhance the organ boundary and reduce the over-segmentation in a non-organ image, yielding more accurate segmentation results. The effectiveness of the proposed method is demonstrated on two datasets. The code is available at: github.com/ZJUGiveLab/UNet-Version
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge