Shurong Chai
A Text-Image Fusion Method with Data Augmentation Capabilities for Referring Medical Image Segmentation
Oct 14, 2025



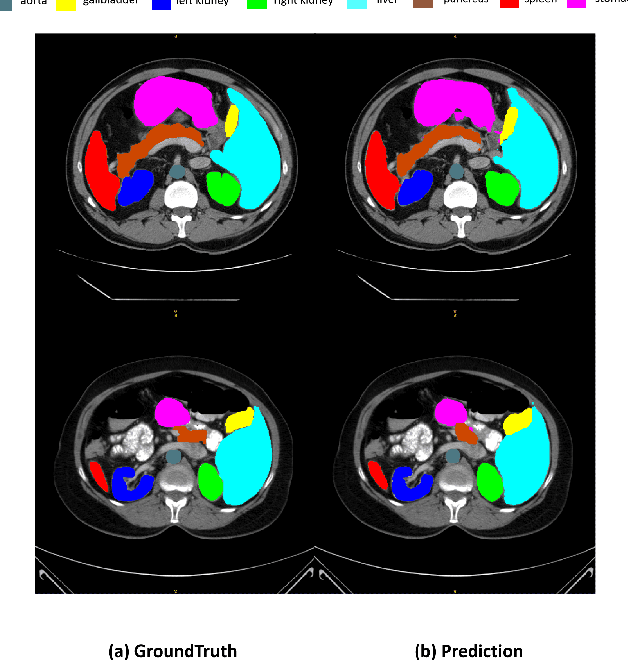
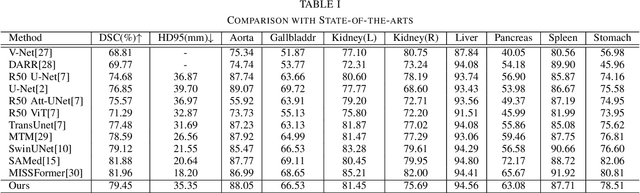
Abstract:Deep learning relies heavily on data augmentation to mitigate limited data, especially in medical imaging. Recent multimodal learning integrates text and images for segmentation, known as referring or text-guided image segmentation. However, common augmentations like rotation and flipping disrupt spatial alignment between image and text, weakening performance. To address this, we propose an early fusion framework that combines text and visual features before augmentation, preserving spatial consistency. We also design a lightweight generator that projects text embeddings into visual space, bridging semantic gaps. Visualization of generated pseudo-images shows accurate region localization. Our method is evaluated on three medical imaging tasks and four segmentation frameworks, achieving state-of-the-art results. Code is publicly available on GitHub: https://github.com/11yxk/MedSeg_EarlyFusion.
Enhancing Depression Detection with Chain-of-Thought Prompting: From Emotion to Reasoning Using Large Language Models
Feb 09, 2025Abstract:Depression is one of the leading causes of disability worldwide, posing a severe burden on individuals, healthcare systems, and society at large. Recent advancements in Large Language Models (LLMs) have shown promise in addressing mental health challenges, including the detection of depression through text-based analysis. However, current LLM-based methods often struggle with nuanced symptom identification and lack a transparent, step-by-step reasoning process, making it difficult to accurately classify and explain mental health conditions. To address these challenges, we propose a Chain-of-Thought Prompting approach that enhances both the performance and interpretability of LLM-based depression detection. Our method breaks down the detection process into four stages: (1) sentiment analysis, (2) binary depression classification, (3) identification of underlying causes, and (4) assessment of severity. By guiding the model through these structured reasoning steps, we improve interpretability and reduce the risk of overlooking subtle clinical indicators. We validate our method on the E-DAIC dataset, where we test multiple state-of-the-art large language models. Experimental results indicate that our Chain-of-Thought Prompting technique yields superior performance in both classification accuracy and the granularity of diagnostic insights, compared to baseline approaches.
Cross-modality Attention Adapter: A Glioma Segmentation Fine-tuning Method for SAM Using Multimodal Brain MR Images
Jul 03, 2023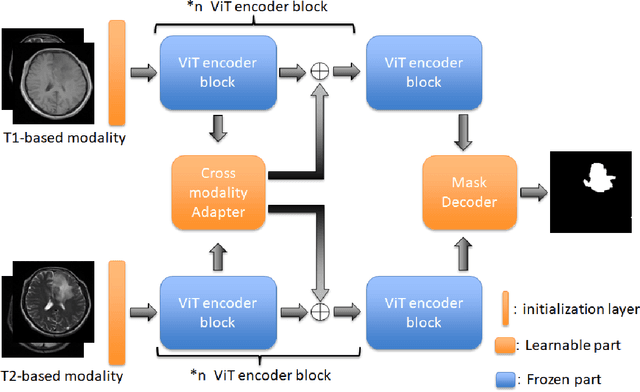
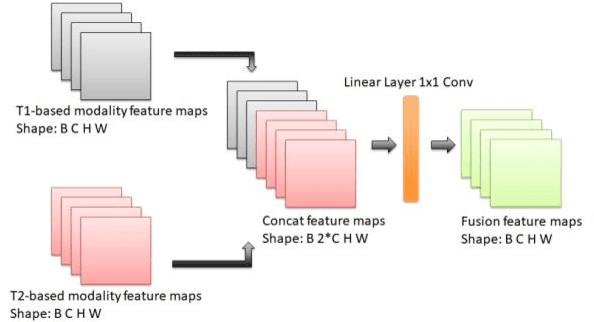

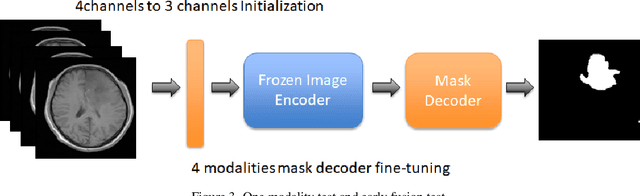
Abstract:According to the 2021 World Health Organization (WHO) Classification scheme for gliomas, glioma segmentation is a very important basis for diagnosis and genotype prediction. In general, 3D multimodal brain MRI is an effective diagnostic tool. In the past decade, there has been an increase in the use of machine learning, particularly deep learning, for medical images processing. Thanks to the development of foundation models, models pre-trained with large-scale datasets have achieved better results on a variety of tasks. However, for medical images with small dataset sizes, deep learning methods struggle to achieve better results on real-world image datasets. In this paper, we propose a cross-modality attention adapter based on multimodal fusion to fine-tune the foundation model to accomplish the task of glioma segmentation in multimodal MRI brain images with better results. The effectiveness of the proposed method is validated via our private glioma data set from the First Affiliated Hospital of Zhengzhou University (FHZU) in Zhengzhou, China. Our proposed method is superior to current state-of-the-art methods with a Dice of 88.38% and Hausdorff distance of 10.64, thereby exhibiting a 4% increase in Dice to segment the glioma region for glioma treatment.
Ladder Fine-tuning approach for SAM integrating complementary network
Jun 22, 2023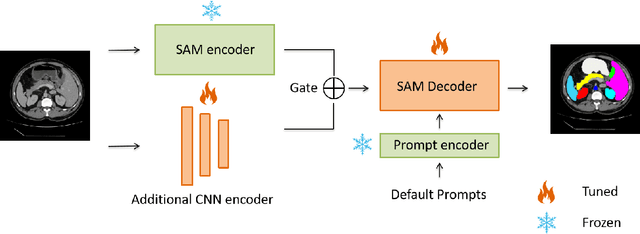

Abstract:Recently, foundation models have been introduced demonstrating various tasks in the field of computer vision. These models such as Segment Anything Model (SAM) are generalized models trained using huge datasets. Currently, ongoing research focuses on exploring the effective utilization of these generalized models for specific domains, such as medical imaging. However, in medical imaging, the lack of training samples due to privacy concerns and other factors presents a major challenge for applying these generalized models to medical image segmentation task. To address this issue, the effective fine tuning of these models is crucial to ensure their optimal utilization. In this study, we propose to combine a complementary Convolutional Neural Network (CNN) along with the standard SAM network for medical image segmentation. To reduce the burden of fine tuning large foundation model and implement cost-efficient trainnig scheme, we focus only on fine-tuning the additional CNN network and SAM decoder part. This strategy significantly reduces trainnig time and achieves competitive results on publicly available dataset. The code is available at https://github.com/11yxk/SAM-LST.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge